Abstract
Introduction
The metabotropic glutamate receptor 4 (mGlu4, GRM4) exhibits significant expression within the central nervous system (CNS) and has been implicated to be correlated with a poor prognosis.
Objective
This study was aimed to elucidate the relationship between the expression profile of GRM4 and the prognosis of glioma patients.
Methods
RNA-sequencing datasets from The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and China Glioma Genome Atlas (CGGA) repositories were used to evaluate the potential relationship. The value of clinical prognostic about GRM4 was assessed using clinical survival data from CGGA and TCGA. The GEPIA database was used to select genes like GRM4. PPI network was constructed by the database of (STRING), GO and KEGG analyses were performed. TargetScan, TarBase, miRDB, and starBase were used to explore miRNAs that could regulate GRM4 expression. EWAS Data Hub, MethSurv, and MEXPRESS were used for the analysis and relationship between DNA methylation and GRM4 expression and prognosis in glioma. TIMER2.0 and CAMOIP databases were used to assess the association between immune cell infiltration and GRM4. Human GBM cell lines were used to validate the function of GRM4.
Results
Our study shows that GRM4 is under expressed among gliomas and accompanied by poorer OS. Multivariate analysis showed that low mRNA expression of GRM4 was an independent factor of prognostic for shorter OS in all glioma patients. MiR-1262 affects the malignant phenotype of gliomas through GRM4. Methylation of DNA plays an important role in the instruction of GRM4 expression, the methylation level of GRM4 in glioma tissue is higher in comparison to normal tissue, and the higher methylation level was accompanied with the worse prognosis. Further analysis showed that GRM4 mRNA expression in GBM linked negatively with common lymphoid progenitor, Macrophage M1, Macrophage, and T cell CD4+ Th2, but not with the tumor purity. Overexpression of GRM4 prevents the migration of human GBM cell lines in vitro.
Conclusion
GRM4 may have a substantial impact on the infiltration of immune cells and serve as a valuable prognostic biomarker in gliomas.
Introduction
Gliomas originate from precursor or glial cells, which subsequently give rise to astrocytoma, ependymoma, oligodendroglioma, and occasionally oligoastrocytoma [Citation1]. These tumors demonstrate an infiltrative growth pattern characterized by the absence of a discernible tumor border. The classification of gliomas by the World Health Organization (WHO) involves four distinct grades. Grade 1 and grade 2 gliomas are categorized as low-grade tumors, while grade 3 and grade 4 gliomas are indicative of high-grade gliomas, also known as HGG [Citation2]. In contrast, glioblastoma (grade 4) is the most common and aggressive glioma. Treatment options depend on type of the cell, location, and grade of the glioma, and are usually a combination of surgery to remove the tumor as safely as possible, radiation therapy (which targets the tumor with a beam of radiation), and chemotherapy (which uses drugs to destroy the cancer cells) [Citation3]. Nevertheless, the median survival for GBM patients remains less than a year from diagnosis [Citation4]. Incidence, however, is increasing due to aging [Citation5,Citation6]. Thus, it is necessary to develop novel therapeutic strategies to advance the prognosis for those with gliomas and meanwhile improve the quality of life of patients. Molecular marker status can guide clinical decision-making and has become an integral part of tumor assessment in modern neuro-oncology practice [Citation7]. The discovery of biomarkers enables a better understanding of these diseases and better selection and development of treatment options.
Metabotropic glutamate (mGlu) receptors are classified as G-protein-coupled receptors (GPCRs) that are primarily expressed in glial cells and neurons. These receptors are located near each synaptic cleft [Citation8]. The MGlu receptors can be categorized into three distinct subfamilies, namely group I (consisting of mGlu1 and mGlu5 receptors), group II (comprising mGlu2 and mGlu3 receptors), and group III (including mGlu4, mGlu6, mGlu7, and mGlu8 receptors), These subfamilies are differentiated based on their sequence homology, second messenger coupling mechanisms, and pharmacological properties) [Citation9]. The expression of GRM4 is notably elevated within the central nervous system (CNS) and it serves various functions in both physiological and pathophysiological processes within the CNS. These functions encompass the facilitation of the learning process, the formation, and retrieval of memories, as well as the potential involvement in cognitive decline [Citation10]. The expression of GRM4 is correlated with unfavorable prognosis in various cancer types, including colorectal cancer, osteosarcoma, breast cancer, and cell carcinoma [Citation11–14]. In addition, glutamate discharge promotes and causes the growth of malignant gliomas [Citation15]. L-Glutamate (Glu) serves as a prominent excitatory neurotransmitter within the central nervous system (CNS) of mammals, mGlu receptors could modulate l-glutamate neurotransmission, and targeting the mGlu4 receptor holds potential benefits in the management of diverse conditions, encompassing various types of cancer, autoimmune diseases, and disorders [Citation8]. However, research on the role of GRM4 in glioma is very inadequate and limited.
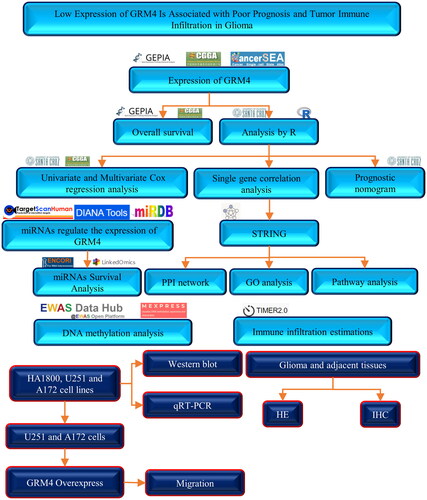
In our study (the flow chart is as follows ), We conducted an evaluation and investigation of the expression of GRM4 in glioma, as reported in four separate cohort studies which include Gene Expression Omnibus (GEO), Chinese Glioma Genome Atlas (CGGA), Genotype-Tissue Expression (TCGA), and The Cancer Genome Atlas (TCGA), in addition to its correlation with patient prognosis. Our experiment indicated and showed that GRM4 was low expressed in glioma. In glioma patients, lower GRM4 expression was associated with a worse OS. Additionally, it was hypothesized that GRM4 plays a role in the modulation of the glutamate receptor signaling pathway, as well as GRM4 expression is associated with miRNAs and DNA methylation. Moreover, our study found that GRM4 expression was not linked to tumor purity, but rather to the invasion of numerous immune cells.
Materials and methods
Data collection and analysis
GEPIA, also known as Gene Expression Profiling Interactive Analysis, is a web-based tool that utilizes data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) projects [Citation16]. The CGGA database is the first complete functional genomics database of Chinese glioma patient cohorts [Citation17]. The GBM microarray data, specifically GSE84465 (Platform: GPL18573) and GSE57872 (Platform: GPL16791), were obtained from the GEO database.
Protein-protein interaction (PPI) network and functional enrichment analysis
The STRING database (http://string-db.org) (version 11.5) was designed to collect all known and anticipated protein interactions, both physical and functional. Enrichment analysis was conducted by STRING using well-known categorization systems like GO and KEGG, knowing that it can offer innovative classification methods based on high flux textual data and clustering algorithms of the correlation network itself [Citation18,Citation19].
Tumor immune estimation resource 2.0 (TIMER 2.0)
(TIMER 2.0) Tumor Immune Estimation Resource 2.0 database (http://timer.cistrome.org/) This system is used to analyze the tumor and its infiltration into immune cells and network resources [Citation20]. The platform enables users to actively investigate the connections between immune infiltration and various factors, such as clinical outcomes, somatic mutations, gene expression, and alterations in somatic copy number.
Cell lines and culture
Obtaining the glioma U251 and A172 cells came from China Wuhan Procell Life Science and Technology Co., LTD., and the healthy human astrocyte HA1800 was purchased from BeNa Culture Collection, Henan, and China. Culture cells according to cell instructions.
qRT-PCR
The isolation of total RNA from cells was performed using a small RNA extraction kit (EZ-10 DNA away, Shengong Biotechnology, Shanghai, China). Subsequently, the obtained RNA was converted into complementary DNA (cDNA) using the Revert Aid First Strand cDNA Synthesis Kit (K1622, US). By employing the established quantitative polymerase chain reaction (qPCR) methodology and utilizing β-Actin as the internal reference, the study aims to accurately measure and compare gene expression levels, the qPCR of GRM4 and β-Actin was performed using NovoStart® SYBR qPCR Super MixPlus on a real-time PCR instrument. Amplification primers (Shanghai General Biology, Shanghai, China) and sequence details are displayed in the . The relative transcription levels of the target genes were determined using the 2ΔΔCT method. The utilization of the average expression was employed to differentiate the cohort with high expression levels from the cohort with low expression levels.
Table 1. Primer sequences.
Western blot analysis
A mixture of RIPA lysate (C500005-0050, Shanghai Shenggong Bioengineering Co., LTD.) and phenylmethyl sulfonyl fluoride (C500005-0050, Shanghai Shenggong Bioengineering Co., LTD.) was used to dissolve the total number of protein obtained by the cells on ice. A BCA protein concentration assay kit (Shanghai Shenggong Bioengineering Co., LTD. C500005-0050) was employed to ascertain the protein concentration of each sample. The protein samples with different concentrations were adjusted to the same concentration by using lysate and 5 × Loading Buffer. Protein samples were isolated by SDS-PAGE electrophoresis and then subsequently transferred to polyvinylidene fluoride (FFP24, Beyotime, China). After the membrane transfer is completed, it is quickly transferred to the pre-prepared rapid sealing fluid for sealing. The sealed PVDF was rinsed with TBST and incubated with the diluted corresponding antibody solution at 4 °C overnight. The primary antibody incubated overnight was recovered the next day, the membrane was washed three times by TBST, and the corresponding diluent of the secondary antibody was incubated for 1h. The second antibody diluent was recovered, rinsed PVDF drops were added with CEL developer, and then scanned in a BIO-RAD gel imager.
Immunohistochemistry
This study of great importance was conducted concerning the principles outlined in the Declaration of Helsinki. The Glioma histopathological sampling and the patients’ clinical diagnosis were down In the Department of Neurosurgery at the First Affiliated Hospital of Xinjiang Medical University. The study has received approval from the research ethics committee of the First Affiliated Hospital of Xinjiang Medical University and all patients consent for publication. Tissue paraffin sections were dewaxed with xylene and rehydrated to repair exposed antigens. 5–10% goat serum (diluted with PBS) the simple was enclosed at room temperature for 10 min, and then incubated with primary antibody working solution at 4 °C overnight. The next day, rinse with PBS, add a suitable amount of biotin-labeled secondary antibody working solution, incubate for 30 min at 37 °C, and rinse again with PBS. Then, an appropriate amount of streptavidin and horseradish peroxidase complex (Biosharp) was added and incubated for 30 min at room temperature, washed with PBS, and color developing agent. Rinse fully with tap water, redye, dehydrate, transparent, and seal.
Statistical analysis
The Statistical analysis has been performed with R (version 3.6.3, accessible at https://www.r-project.org/). We used Cox proportional hazards modeling in both univariate and multivariate studies to quantify and assess the risk of death. Potential confounding variables in this study included age, gender, clinical stage, and treatment. A significance level of p < 0.05 was employed to assess the statistical significance.
Results
GRM4 is downregulated in glioma
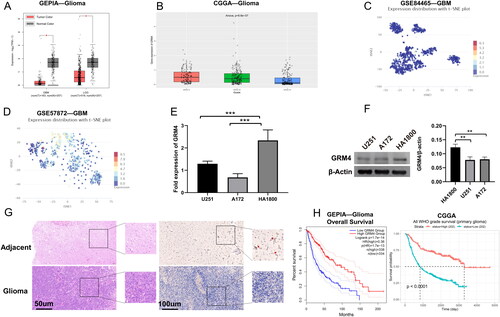
According to the database of GEPIA (http://gepia2.cancer-pku.cn/), GRM4 mRNA appearance was lower in LGG and GBM tissues than in normal tissues (). Based on the CGGA database (http://www.cgga.org.cn/index.jsp), GRM4 mRNA expression was lower in high-grade gliomas (). In addition, based on the CancerSEA database (http://biocc.hrbmu.edu.cn/CancerSEA/), we investigated the expression level of GRM4 mRNA in GSE84465 and GSE57872 datasets [Citation21,Citation22] (). The qPCR analysis and Western blot revealed that GRM4 mRNA and protein were both markedly low-expressed in the human glioma cells (U251, A172) compared to human astrocytes (HA1800, ). Again, we got the same results in glioma tissue and paratumoral tissue. (). Collectively, these results strongly indicate that GRM4 exhibits downregulation in human glioma.
Figure 2. The low expression of GRM4 in glioma is associated with prognosis. (A) According to the GEPIA database, the mRNA expression level of GRM4 in glioma tissues exhibited a statistically significant decrease compared to normal tissues. (B) Based on the data obtained from the CGGA database, the mRNA expression of GRM4 was found to be reduced in high-grade gliomas (C-D)GRM4 mRNA expression based on CancerSEA database GSE84465 and GSE57872 datasets. T-SNE is utilized to depict the cellular distribution, where each point corresponds to a distinct cell. The color of each point signifies the expression level of GRM4 within the respective cell. (E-F) expression of the GRM4 in the pertinent groups was evaluated through the utilization of qPCR and Western blot. All the data are shown as the mean ± SD (three independent experiments). Sig: ns p ≥ 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. (G) HE staining and IHC representative images of glioma and adjacent tissues. (H) Overall survival Kaplan-Meier analysis revealed that glioma patients with low GRM4 mRNA expression had a shorter OS than patients with high GRM4 levels.
The downregulation of GRM4 in glioma is correlated with poor patient survival
Disease prognostication is an integral component of oncology and medicine. To understand completely the correlation between GRM4 mRNA expression levels and prognosis in glioma, we again consulted GEPIA2 and CGGA databases. The Kaplan-Meier survival curve shows that patients with low GRM4 expression had significantly shorter overall survival times than those with high GRM4 expression ().
Additionally, we examined the relative risks that GRM4 indicated for the prognosis of glioma. Cox-regression analysis is mostly used for indicating and determining if GRM4 possibly serves as a factor risk. As shown in , The death risk of the glioma patients was increased significantly and associated with the low expression of GRM4. (p < 0.001) in the comparison of the high GRM4 expression by the univariate Cox regression analyses. The Multivariate Cox regression analysis showed that GRM4 might be a factor for predicting poor survival when GRM4 low expression (p = 0.003), high grade (p < 0.001), IDH wt (p < 0.001), and age (p < 0.001) were included (). These results show a significant relationship between expressions of GRM4 with the glioma prognosis.
Figure 3. GRM4 In gliomas: prognostic, functional, and pathway enrichment analyses (A-B) univariate and multivariate analyses. HR, hazard ratio. (C) Overall survival predictions were based on the nomogram of the glioma cohort (TCGA-GBMLGG). (D) A network formed by the co-expression genes and GRM4. (E) Significant gene Ontology terms of GRM4, including biological processes, molecular function, and cellular components (BP, MF, CC). (F) GMR4 and other associated significant pathways.
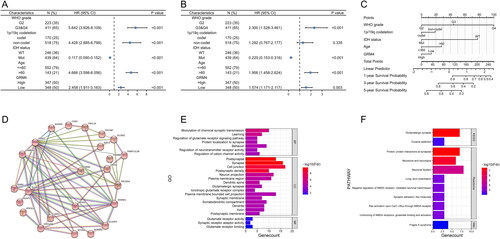
With the capability of producing the numerical probability of individual and clinical events incidents throughout diverse diverse prognostic integrity and finding variables determinant, Nomograms would justify the desires for clinically and biologically integrated models and our ambitions toward personalized medicine [Citation23]. A nomogram was generated on grade (WHO), 1p/19q codeletion in this study, the status of IDH, age, and GRM4 for prediction of the probability of 1-year, 3-year, and 5-year OS. Simultaneously, the C index was computed to be 0.847 with a 95% confidence interval (CI) of 0.836–0.859. Several variables were assessed and scored according to their respective contributions to the risk of survival, as depicted in . Collectively; these results indicate that low expression of GRM4 in glioma patients correlates with poor survival.
Correlation and enrichment analyses
For GRM4 function prediction, including their associated pathways, we downloaded 300 genes similar to GRM4 from the GEPIA database. Then we constructed the PPI network using the STRING database and selected the cluster containing GRM4 for GO analysis and path analysis (). The results of functional enrichment and Gene Ontology (GO) analysis indicate that GRM4 is predominantly linked to gene terms related to synapses, plus Modulation of chemical synaptic transmission, Learning, Behavior, Protein localization to synapse, Glutamate receptor signaling pathways regulation, neurotransmitter receptor activity regulation, and regulation of cation channel activity, Synapse, Postsynapse, Postsynaptic density, Cell junction, Neuron projection, Dendritic spine, Plasma membrane region, Glutamatergic synapse, Ionotropic glutamate receptor complex, Synaptic membrane, Plasma membrane-bounded cell projection, Somatodendritic compartment, Dendrite, Axon, Postsynaptic membrane, Glutamate receptor activity, Synaptic receptor adaptor activity, Glutamate receptor binding (). Additionally, pathway analysis showed that the GRM4 and its co-expression genes were enriched and interacted with in Glutamatergic synapse, Cocaine addiction, Neuronal System, Neurexins and neuroligins, Negative regulation of NMDA receptor-mediated neuronal transmission, Ras activation upon Ca2+ influx through NMDA receptor, Long-term potentiation, Synaptic adhesion-like molecules, Protein-protein interactions at synapses, Unblocking of NMDA receptors, glutamate binding and activation, and Fragile X syndrome (). The results of this study suggest a connection between GRM4 and brain activity, particularly in terms of glutamatergic neurotransmission.
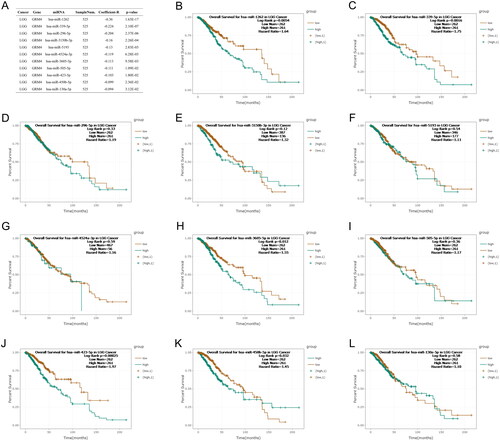
miRNAs regulate the expression of the GRM4 gene in glioma
A growing number of studies have shown that miRNAs are crucial in cancer biology. MicroRNAs (miRNAs) exert regulatory control on gene expression at the post-transcriptional level by primarily interacting with the 3′ untranslated region (3′UTR) of target mRNA molecules [Citation24]. It was reported that the direct binding of miR-328-3p and miR-370-3p to the 3′ untranslated region (UTR) of GRM4 leads to the downregulation of GRM4 expression, thereby facilitating cell proliferation, migration, and invasion in breast cancer [Citation13]. Here, we used TargetScan (http://www.targetscan.org/), TarBase (https://dianalab.e-ce.uth.gr/), and miRDB (http://www.mirdb.org/) to identify miRNAs that might regulate GRM4 expression [Citation25–27]. The results show that hsa-miR-1262, hsa-miR-339-5p, hsa-miR-296-5p, hsa-miR-3150b-3p, hsa-miR-5193, hsa-miR-4524a-3p, hsa-miR-3605-5p, hsa-miR-505-5p, hsa-miR-423-5p, hsa-miR-450b-5p, and hsa-miR-130a-5p might regulate GRM4 expression (). Next, We utilized the starBase platform (https://starbase.sysu.edu.cn/) to assess the correlation between miRNA-GRM4 expression and analyze the relationship between miRNAs expression and prognosis in LGG [Citation28]. The results show that patients with hsa-miR-1262, has-miR-3605, and has-miR-423 high expression of LGG had a worse prognosis (). Subsequently, we examined the expression of hsa-miR-1262, has-miR-3605, and has-miR-423 in TCGA_GBM&LGG, and their relationship with prognosis and GRM4 through LinkedOmics [Citation29] (http://www.linkedomics.org/ Supplementary Figure 1). Together, our analysis showed that hsa-miR-1262 may as the most plausible regulator of GRM4 in glioma.
GRM4 expression is associated with DNA methylation
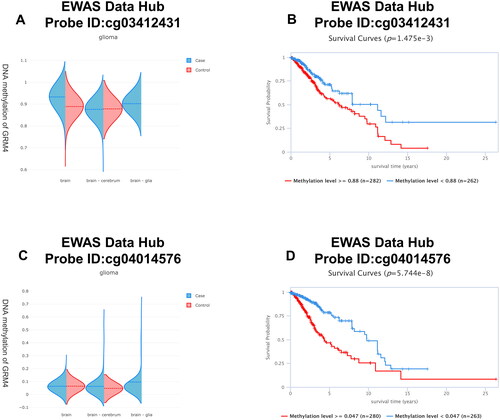
DNA methylation is a prevalent epigenetic occurrence that plays a critical role in the development of cancer. The methylation patterns typically observed in cells of a tumor exhibit a recurring disruption, characterized by a combination of global hypomethylation and region-specific hypermethylation. When a tumor suppressor gene’s promoter is hypermethylated, it results in the silencing of the gene and confers a growth advantage to the cell, similar to mutations or deletions [Citation30]. Hence, we investigated the connection between GRM4 expression levels and DNA methylation in gliomas. Based on the EWAS Data Hub (https://ngdc.cncb.ac.cn/ewas/datahub), the methylation level of GRM4 in glioma tissue is higher compared to normal tissue, and the higher the methylation level, the worse the prognosis ().
Figure 5. GRM4 methylation in glioma. (A, C) The methylation level of the GRM4 gene in glioma tissue exhibits a higher magnitude compared to that observed in normal tissue. (B, D) Patients exhibiting high levels of GRM4 methylation demonstrated a significantly inferior overall survival outcome in comparison to patients with low levels of GRM4 methylation.
The MethSurv tool was employed to examine the prognostic significance of individual CpG sites to DNA methylation levels of GRM4 [Citation31]. The MethSurv analysis yielded a total of 50 CpG sites exhibiting methylation, with cg00143321 and cg05745748 displaying the most prominent levels of DNA methylation (). Five CpG sites’ levels of methylation, namely cg00143321, cg20490197, cg00070899, cg15884992, and cg20151098, were related with prognosis (p < 0.05, ). T those Patients with high GRM4 methylation of CpG sites had worse overall survival compared to patients with low GRM4 methylation. MEXPRESS is a web-based tool utilized for the analysis of clinical data, DNA methylation, and visualization of TCGA expression of the Gene and the relationship among them [Citation32,Citation33]. Using MEXPRESS, we explored the correlation of the DNA methylation data with GRM4 gene expression in LGG and GBM (Supplementary Figure 2–3). Together, these data indicated that DNA methylation is essential to controlling GRM4 expression in gliomas.
Figure 6. DNA methylation levels in the GRM4 gene are associated with the prognosis of GBM patients.
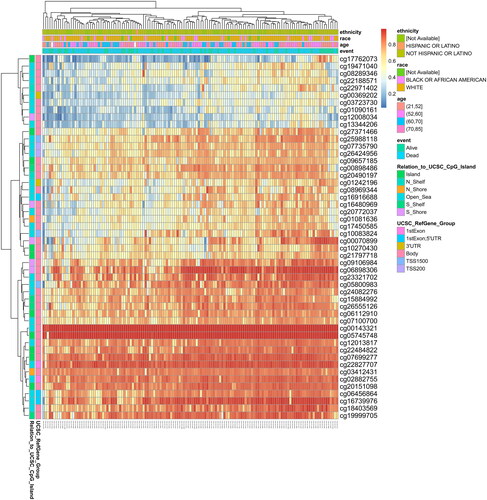
Table 2. Effects of methylation levels in the CpG sites of the GRM4 gene on the prognosis of GBM patients.
GRM4 overexpression inhibited GBM cell migration in vitro
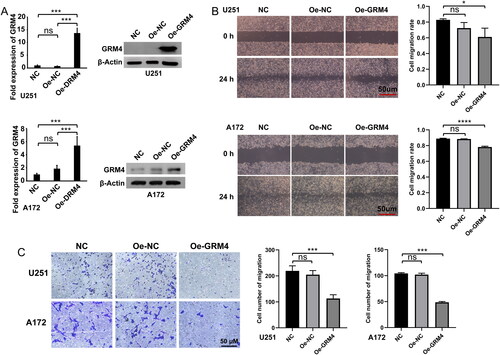
To further investigate the function of GRM4, U251, and A172 cells were transduced by lentivirus to overexpress GRM4. The qPCR and WB assays showed that GRM4 expression was significantly increased in both U251 and A172 cells overexpressing lentiviral transduction compared with negative controls (). To investigate the impact of GRM4 overexpression upon the malignant phenotype of GBM cells, we conducted in vitro cell experiments. The results of the wound healing and transwell cell migration assay showed that the overexpression of GRM4 had a significant inhibitory effect on the migratory capacity of U251 and A172 cells ().
Figure 7. GRM4 overexpression inhibited GBM cell migration in vitro. (A) The expression of GRM4 in U251 and A172 glioma cells was detected through the utilization of qPCR and Western blot analysis after lentiviral transduction. (B) Wound healing assay images and statistical results, scale = 50 μm. (C)Transwell assay images and statistical results, scale = 50 μm.
GRM4 expression is associated with immune cell infiltration
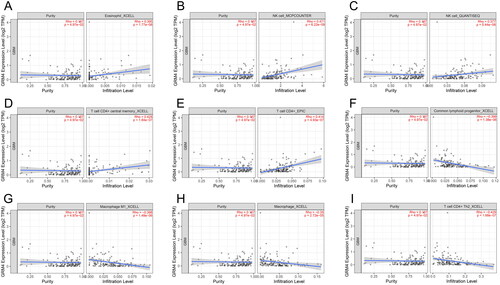
Tumor-infiltrating immune inflammatory cells assume a crucial role in the progression of tumorigenesis and the development of malignancy [Citation34]. To understand the correlation of immune cell infiltration with GRM4, we used TIMER2.0 (http://timer.cistrome.org/) [Citation20,Citation35,Citation36]. According to TIMER2.0 analysis, GRM4 mRNA expression in GBM was positively correlated with Eosinophil_XCELL (R = 0.395, p = 1.77E-06), NK cell_MCPCOUNTER (R = 0.471, p = 3.09E-07), NK cell_QUANTISEQ (R = 0.377, p = 5.44E-06), T cell CD4+ central memory_XCELL (R = 0.428, p = 1.84E-07), and T cell CD4+_EPIC (R = 0.414, p = 1.84E-07), and was negatively linked with Common lymphoid progenitor_XCELL (R= −0.399, p = 1.38E-06), Macrophage M1_XCELL (R = −0.398, p = 1.48E-06), Macrophage_XCELL (R = −0.35, p = 2.72E-05), T cell CD4+ Th2_XCELL (R = −0.429, p = 1.68E-07), but not with the tumor purity (R = 0.167, p = 4.97E-02) (). The correlation between GRM4 mRNA expression and immune cells in LGG is shown in Supplementary Table 1. This indicates that low expression of GRM4 has a strong association with immune cell infiltration gliomas.
Discussion
Most mGlu receptors are found all over the brain and are widely expressed, mGlu3 and mGlu5 were present in glial cells, while mGlu6 was restricted to the retina [Citation9]. In the adult cerebellar cortex, the expression levels of GRM1 and GRM4 are predominantly high in granule cells, whereas the expression levels of GRM3 and GRM7 are considerably lower. Additionally, transcripts for the receptor specific to the retina are also present [Citation37]. Under such pathological conditions it has been suggested that mGluR activation is involved in neurodegenerative processes [Citation38], Reduced mGluR4 gene expression is caused by neuronal apoptosis and is linked to it. MGluR4 selective activity also protects against excitotoxication-induced neuronal death [Citation39]. Our study shows that GRM4 is underexpressed in gliomas and is related to poorer OS, overexpression of GRM4 inhibited glioma cell migration. Multivariate analysis indicated that low mRNA expression of GRM4 was an independent prognostic factor for shorter OS in all glioma patients. Meanwhile, GO and KEGG analysis indicated that GRM4 is associated with brain function and glutamatergic neurotransmission. The results of this study show that GRM4 exhibits promise as a potential biomarker for prognosticating the outcomes of patients with tumors.
The dysregulation of microRNA (miRNA) expression has been implicated in the pathogenesis of several types of cancer, acting as tumor suppressors and oncogenes, controlling mRNA expression, and affecting malignant phenotypes such as apoptosis, angiogenesis, invasion/metastasis, and proliferation. Prior research has demonstrated that miRNAs regulate the expression of GRM4 in various diseases[Citation13,Citation40,Citation41]. our analysis showed that hsa-miR-1262 may as the most plausible regulator of GRM4 in glioma. In addition, miR-1262 could affect the prognosis of gastric cardia adenocarcinoma, breast cancer, and lung cancer patients [Citation42–44]. Therefore, we speculate that miR-1262 affects the malignant phenotype of gliomas through GRM4.
Typically, changes in gene expression occur through methylation or demethylation. The differential regulation of histone methylation associated with open chromatin at corresponding promoters is indicative of region-specific variations in the presentation and expression of metabotropic glutamate receptor genes in the human brain [Citation45]. The cerebellar cortex exhibits elevated GRM4 promoter’s H3K4me2 and H3K4me3 levels, along with increased levels of histone methylation associated with open chromatin [Citation45]. Our research shows that DNA methylation is essential for controlling the expression of GRM4, the methylation level of GRM4 in glioma tissue is higher cooperation to normal tissue, and the prognosis deteriorates as the methylation level increases.
The immune system tremendously adds and contributes to preventing the occurrence of the tumor, progression, and metastasis of tumors and regulating tumor cells’ response to therapy. The expression of mGluR4 was observed in all peripheral dendritic cells (DCs) and specific subtypes of CD4+ T cells, such as Treg cells but not TH17 cells, in mice lacking mGluR4. In these mice, an immune response characterized by the production of IL-17 by TH17 cells resulted in the progression of experimental autoimmune encephalomyelitis (EAE). Conversely, the administration of selective mGluR4 enhancers was capable of inducing a protective response dominated by CD4+ Treg cells [Citation46]. Moreover, preclinical study results show that the activation of metabotropic glutamate receptor 4 (mGlu4) may potentially offer therapeutic benefits in the management of medulloblastoma [Citation47]. GRM4 mRNA expression in GBM Was linked negatively with Common lymphoid progenitor, Macrophage M1, and Macrophage, T cell CD4+ Th2, but not with the tumor purity. In this study, we preliminarily explored the mechanism of low expression of GRM4 in gliomas through bioinformatics methods, including miRNA and DNA methylation, but it was not experimentally validated. In future studies, we will conduct a series of experiments to further verify the regulation of GRM4 expression by miRNA or DNA methylation and its relationship with immune cell infiltration.
In summary, it can be inferred that GRM4 potentially assumes a critical function in the infiltration of immune cells and serves as a significant prognostic biomarker in Glioma.
Author contributions
Hai Fan analyzed the data, contributed reagents/materials/analysis tools, authored and reviewed drafts of the paper, and approved the final draft. Dongming Yan conceived and designed the study, and authored and reviewed drafts of the paper. Xingyue Fang performed the experiments, analyzed the data, and approved the final draft. Liumin Xiao, Mengjie Liang, and Haolin Wu performed data analysis, wrote the manuscript, analyzed the data, and prepared figures and/or tables. Guohua Zhu, Dangmurenjiafu Geng, and Qibing Liu authored or reviewed the draft of the paper and approved the final draft. In preparing this work, the author did not use any artificial intelligence AIDS. The authors confirm that the data supporting the findings of this study are available within the article [and/or its supplementary materials. The authors declare no conflict of interest and consent to publication.
Supplemental Material
Download TIFF Image (2.2 MB)Supplemental Material
Download PDF (6.1 MB)Supplemental Material
Download TIFF Image (17.2 MB)Acknowledgments
The authors thank the GEPIA, CGGA, TCGA, TargetScan, TarBase, miRDB, STRING, EWAS, MethSurv, and MEXPRESS databases for providing their platforms and contributors for their valuable data sets.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Xu S, Tang L, Li X, et al. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476(2020):1–12. doi:10.1016/j.canlet.2020.02.002.
- Louis DN, Perry A, Reifenberger G, et al. The 2016 world health organization classification of tumors of the Central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1.
- Chang HJ, Burke AE, Glass RM. JAMA patient page. Gliomas JAMA. 2010;303(10):1000. doi:10.1001/jama.303.10.1000.
- Gittleman H, Boscia A, Ostrom QT, et al. Survivorship in adults with malignant brain and other Central nervous system tumor from 2000-2014. Neuro Oncol. 2018;20(suppl_7):vii6–vii16. doi:10.1093/neuonc/noy090.
- Korja M, Raj R, Seppä K, et al. Glioblastoma survival is improving despite increasing incidence rates: a nationwide study between 2000 and 2013 in Finland. Neuro Oncol. 2019;21(3):370–379. doi:10.1093/neuonc/noy164.
- Miranda-Filho A, Pineros M, Soerjomataram I, et al. Cancers of the brain and CNS: global patterns and trends in incidence. Neuro Oncol. 2017;19(2):270–280.
- Weller M, Pfister SM, Wick W, et al. Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol. 2013;14(9):e370–e9. doi:10.1016/S1470-2045(13)70168-2.
- Julio-Pieper M, Flor PJ, Dinan TG, et al. Exciting times beyond the brain: metabotropic glutamate receptors in peripheral and non-neural tissues. Pharmacol Rev. 2011;63(1):35–58. doi:10.1124/pr.110.004036.
- Suh Y H, Chang K, Roche KW. Metabotropic glutamate receptor trafficking. Mol Cell Neurosci. 2018;91:10–24. doi:10.1016/j.mcn.2018.03.014.
- Wan Z, Sun R, Yang-Wuyue LIU, et al. Targeting metabotropic glutamate receptor 4 for cancer immunotherapy. Sci Adv. 2021;7(50):eabj4226. doi:10.1126/sciadv.abj4226.
- Chang HJ, Yoo BC, Lim S-B, et al. Metabotropic glutamate receptor 4 expression in colorectal carcinoma and its prognostic significance. Clin Cancer Res. 2005;11(9):3288–3295. doi:10.1158/1078-0432.CCR-04-1912.
- Kansara MAYA, Thomson K, Pang P, et al. Infiltrating myeloid cells drive osteosarcoma progression via GRM4 regulation of IL23. Cancer Discov. 2019;9(11):1511–1519. doi:10.1158/2159-8290.CD-19-0154.
- Xiao B, Chen D, Zhou Q, et al. Glutamate metabotropic receptor 4 (GRM4) inhibits cell proliferation, migration and invasion in breast cancer and is regulated by miR-328-3p and miR-370-3p. BMC Cancer. 2019;19(1):891. doi:10.1186/s12885-019-6068-4.
- Huang CY, Hsueh YM, Chen LC, et al. Clinical significance of glutamate metabotropic receptors in renal cell carcinoma risk and survival. Cancer Med. 2018;7(12):6104–6111. doi:10.1002/cam4.1901.
- Takano T, Lin JH, Arcuino G, et al. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7(9):1010–1015. doi:10.1038/nm0901-1010.
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247.
- Zhao Z, Zhang KN, Wang Q, et al. Chinese glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from chinese glioma patients. Genomics Proteomics Bioinformatics. 2021;19(1):1–12. doi:10.1016/j.gpb.2020.10.005.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131.
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi:10.1093/nar/gkaa1074.
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407.
- Darmanis S, Sloan SA, Croote D, et al. Single-Cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. doi:10.1016/j.celrep.2017.10.030.
- Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi:10.1126/science.1254257.
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173–e80. doi:10.1016/S1470-2045(14)71116-7.
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4(1):199–227. (doi:10.1146/annurev.pathol.4.110807.092222.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035.
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1):D239–D245. doi:10.1093/nar/gkx1141.
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. [Nucleic Acids Res. 2020;48(D1):D127–D131. doi:10.1093/nar/gkz757.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi:10.1093/nar/gkt1248.
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi:10.1093/nar/gkx1090.
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21(3):461–467. doi:10.1093/carcin/21.3.461.
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi:10.2217/epi-2017-0118.
- Koch A, DE Meyer T, Jeschke J, et al. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16(1):636. (doi:10.1186/s12864-015-1847-z.
- Koch A, Jeschke J, VAN Criekinge W, et al. MEXPRESS update 2019. Nucleic Acids Res. 2019;47(W1):W561–W565. doi:10.1093/nar/gkz445.
- Yan D, Li W, Liu Q, et al. Advances in immune microenvironment and immunotherapy of isocitrate dehydrogenase mutated glioma. Front Immunol. 2022;13(7):914618. doi:10.3389/fimmu.2022.914618.
- Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7.
- Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-Infiltrating immune cells. Cancer Res. 2017;77(21):e108-e10–e110. doi:10.1158/0008-5472.CAN-17-0307.
- Berthele A, Platzer S, Laurie DJ, et al. Expression of metabotropic glutamate receptor subtype mRNA (mGluR1-8) in human cerebellum. Neuroreport. 1999;10(18):3861–3867. doi:10.1097/00001756-199912160-00026.
- Miller S, Kesslak JP, Romano C, et al. Roles of metabotropic glutamate receptors in brain plasticity and pathology. Ann N Y Acad Sci. 1995;757(1):460–474. (doi:10.1111/j.1749-6632.1995.tb17506.x.
- Borodezt K, D’mello SR. Decreased expression of the metabotropic glutamate receptor-4 gene is associated with neuronal apoptosis. J. Neurosci. Res. 1998;53(5):531–541. doi:10.1002/(SICI)1097-4547(19980901)53:5<531::AID-JNR3>3.0.CO;2-A.
- Lopez JP, Lim R, Cruceanu C, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med. 2014;20(7):764–768. doi:10.1038/nm.3582.
- Li J, Meng H, Cao W, et al. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci Lett. 2015;606:167–172. doi:10.1016/j.neulet.2015.08.038.
- Zheng Y, Xie M, Zhang N, et al. miR-1262 suppresses gastric cardia adenocarcinoma via targeting oncogene ULK1. J Cancer. 2021;12(4):1231–1239. doi:10.7150/jca.46971.
- Li L, Qu WH, Ma HP, et al. LRP8, modulated by miR-1262, promotes tumour progression and forecasts the prognosis of patients in breast cancer. Arch Physiol Biochem. 2022;128(3):657–665. doi:10.1080/13813455.2020.1716019.
- Xie K, Chen M, Zhu M, et al. A polymorphism in miR-1262 regulatory region confers the risk of lung cancer in chinese population. Int J Cancer. 2017;141(5):958–966. doi:10.1002/ijc.30788.
- Stadler F, Kolb G, Rubusch L, et al. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem. 2005;94(2):324–336. doi:10.1111/j.1471-4159.2005.03190.x.
- Fallarino F, Volpi C, Fazio F, et al. Metabotropic glutamate receptor-4 modulates adaptive immunity and restrains neuroinflammation. Nat Med. 2010;16(8):897–902. doi:10.1038/nm.2183.
- Iacovelli L, Arcella A, Battaglia G, et al. Pharmacological activation of mGlu4 metabotropic glutamate receptors inhibits the growth of medulloblastomas. J Neurosci. 2006;26(32):8388–8397. doi:10.1523/JNEUROSCI.2285-06.2006.