Abstract
Complex respiratory diseases are a significant challenge for the livestock industry worldwide. These diseases considerably impact animal health and welfare and cause severe economic losses. One of the first lines of pathogen defense combines the respiratory tract mucus, a highly viscous material primarily composed of mucins, and a thriving multi-kingdom microbial ecosystem. The microbiome-mucin interplay protects from unwanted substances and organisms, but its dysfunction may enable pathogenic infections and the onset of respiratory disease. Emerging evidence also shows that noncoding regulatory RNAs might modulate the structure and function of the microbiome-mucin relationship. This opinion paper unearths the current understanding of the triangular relationship between mucins, the microbiome, and noncoding RNAs in the context of respiratory infections in animals of veterinary interest. There is a need to look at these molecular underpinnings that dictate distinct health and disease outcomes to implement effective prevention, surveillance, and timely intervention strategies tailored to the different epidemiological contexts.
Introduction
Complex respiratory diseases entail multifactorial processes whose mechanisms are still not fully understood and are a significant problem for animal health and welfare, particularly in intensive systems. Moreover, these diseases can cause economic losses due to reduced productivity, increased morbidity, premature mortality, treatment costs, and severe consequences for public health and the environment (Ericsson et al. Citation2016; Bond et al. Citation2017; Oladunni et al. Citation2019; Ericsson et al. Citation2020; Mach et al. Citation2021). There is a growing awareness in livestock physiopathology that the virulence of infectious agents can be affected by multispecies synergic interactions (Kuiken et al. Citation2005; Holt et al. Citation2011; Ericsson et al. Citation2016; Bond et al. Citation2017; Oladunni et al. Citation2019; Ericsson et al. Citation2020; Blakebrough-Hall et al. Citation2020). Consequently, a central finding of disease complexes involves interactions among holobionts (the host and the many other microorganisms living in or around it (Simon et al. Citation2019)) and multiple etiological agents (Mach et al. Citation2021).
New evidence shows that the airway microbiota, defined as the complex community of microorganisms living in the respiratory tract, including bacteria, eukaryotes, and archaea (Zeineldin et al. Citation2019), might act as a gatekeeper that provides resistance to infection on the mucosal surface (Man et al. Citation2017; Li et al. Citation2019). Under normal physiological conditions, commensal microorganisms maintain a mutualistic relationship with the host by regulating the airway’s innate and adaptive immune functions (Mach et al. Citation2021). Classifying healthy versus diseased animals based on their respiratory tract microbiomes has been done successfully for ruminants (Holman et al. Citation2015; Nicola et al. Citation2017; Gaeta et al. Citation2017; Zeineldin et al. Citation2017; Timsit et al. Citation2017; McMullen et al. Citation2019; Zeineldin et al. Citation2020; McMullen et al. Citation2020; Chai et al. Citation2022; Mariadassou et al. Citation2023), pigs (Correa-Fiz et al. Citation2016; Wang et al. Citation2018; Correa-Fiz et al. Citation2019; Mahmmod et al. Citation2020), horses (UCVM Class of 2019, 2020), and chickens (Yitbarek et al. Citation2018; Ngunjiri et al. Citation2019; Yitbarek et al. Citation2019). However, mucus is an often forgotten aspect of the complex respiratory system in animals.
While mucus has historically been viewed as a simple physical barrier, recent work has suggested that mucins, the major gel-forming components of mucus, have many structural and functional roles in the respiratory system (Rose and Voynow Citation2006; Thornton et al. Citation2008; Thai et al. Citation2008; Ridley and Thornton Citation2018; Atanasova and Reznikov Citation2019). Mucins have a high molecular weight and are made of glycosylated proteins with hundreds of branching chain carbohydrates (O-linked glycans). The carbohydrate structures found in mucins and their arrangements, particularly within glycoproteins, comprise the glycome (Kunej Citation2019, Shipunov and Kupaev Citation2022).
The vast diversity of mucin glycome patterns makes them ideal for encoding biological information with a higher specificity (Shipunov and Kupaev Citation2022). First, they provide a physical and chemical barrier that traps and clears inhaled particles and neutralizes toxins, allergens, pollutants, and pathogens (Rose and Voynow Citation2006; Schnaar Citation2015; Atanasova and Reznikov Citation2019; Reily et al. Citation2019; Qin and Mahal Citation2021; Brazil and Parkos Citation2022). Second, the mucin glycome has also been shown to have immune-modulating properties (Hansson Citation2019). Third, the mucin glycome affects the respiratory microbiota composition and functions as an environmental niche and a carbon and nitrogen source (Rose and Voynow Citation2006; Bergstrom et al. Citation2017; Chatterjee et al. Citation2020). Reciprocally, the respiratory tract microbiota’s composition and activity influence mucins’ production and structure, consequently affecting the host’s ability to fight against pathogenic infections (Pérez-Cobas et al. Citation2023). Any disruption in mucin structure, quantity, or function and the glycosylation pattern of their glycans can lead to dysbiosis and potentially increase the risk of respiratory infections in livestock (Miao et al. Citation2022).
Importantly, recent studies show that noncoding RNAs (ncRNAs) tightly regulate mucin production, secretion, and glycosylation at the genomic level (Skovgaard et al. Citation2013; Agrawal et al. Citation2014; Fleming and Miller Citation2019; Zhang et al. Citation2019). The glycans on mucins are products of multiple glycosyltransferases and glycosidases working in a coordinated manner (Thu and Mahal Citation2020). Additionally, there are variations in glycan distribution in the respiratory tract between species and different locations within the host (Wallace et al. Citation2021). One type of ncRNAs implicated in mucin glycome regulation are microRNAs (miRNAs) (Agrawal et al. Citation2014; Thu and Mahal Citation2020; Jame-Chenarboo et al. Citation2022), which are small (∼22 nucleotides) RNA molecules that bind to messenger RNAs (mRNAs) and regulate their translation and stability into proteins. Several miRNAs have been shown to regulate the glycan biosynthetic enzymes and regulate the expression of mucin genes in the respiratory tract of humans, including miR-34b/c (Li et al. Citation2021), miR-146a (Zhong et al. Citation2011), miR-378 (Skrzypek et al. Citation2013), and miR-141 (Siddiqui et al. Citation2021), but also in livestock (Brogaard et al. Citation2018; Skovgaard et al. Citation2013; Timoneda et al. Citation2014; Fleming and Miller Citation2019).
This review aims to give insights into the potential avenues of complex respiratory disease, building on the surge of recent primary research to debate different aspects of the complex and intricate relationships between pathogens, the holobiont, mucins, and their genomic regulation. Underpinning these mechanisms will be crucial for determining how these can be harnessed to develop novel interventions to prevent disease infection and improve animal health and welfare.
Materials and methods
A systematic review and synthesis of relevant qualitative research was conducted according to the requirements established in the preferred reporting items for systematic review and meta-analysis protocols (PRISMA) (the PRISMA-P Group, 2015).
Eligibility criteria and literature search strategy
A systematic and comprehensive search of electronic databases, including Medline database (https://pubmed.ncbi.nlm.nih.gov/), Scopus, ClinicalTrials.gov, Science Direct, Springer Link, Google Scholar, and EMBASE was done from January 2023 to October 2023. The search process was completed using the keywords: ‘mucin respiratory tract’, ‘respiratory complex disease’, ‘microbiota’, ‘pathogens’, ‘livestock’, ‘non-coding RNAs’, and ‘animals of veterinary interest’. The search was not restricted to the type of study (e.g. case-control, prospective cohort studies, randomized control trial, before-and-after study, cross-over randomized control trial), sample size, age, sex, breed, geographic localization, or species. However, editorials, systematic and literature reviews, and letters to the editors were excluded. Our analysis did not include studies focusing on specific medical conditions, treatments, or demographics. We only considered peer-reviewed and original research studies published from 2000 forward and in English only. All papers were exported to the reference database Mendeley.
Data extraction
Complete copies of citations coded as potentially relevant were obtained, and those meeting the inclusion criteria were read in detail, and data were extracted. The author pulled information about the species, study aims, population and sample size, experimental design and duration of follow-up, individual characteristics, changes in the microbiota composition, mucin characterization, and association or not with ncRNAs. The primary outcome was the microbiota profile, aberrant changes in the mucin composition, abundance, or structure, and other relevant outcomes related to ncRNAs and microbiota-mucin interactions. If eligibility could be determined, the entire article was retrieved.
Data synthesis
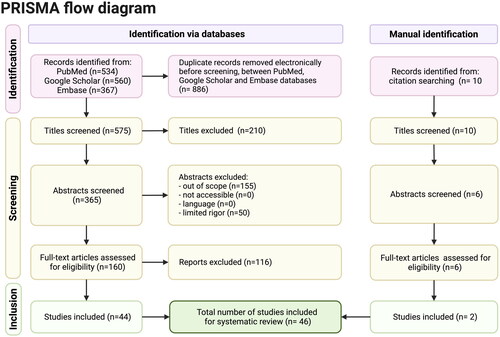
The search conducted on January 2023 resulted in 1461 articles, most of which were cross-matched between databases. Of the remaining and unique 575 articles, we observed the following list of key term combinations: respiratory complex disease, microbiota, and animal = 156; airway mucins and respiratory disease =195; mucins, respiratory illness and sialic acid = 76; airway mucins and respiratory microbiota = 30; airway mucins, respiratory microbiota and infection = 17; non-coding RNAs and mucins = 91; and the ncRNAs, mucins, and livestock = 10. After screening by title and abstract, 160 papers were identified as meeting the inclusion criteria. Following a full-text article assessment for eligibility, 116 more papers were excluded, leaving 46 experimental studies on farm and companion animals eligible for inclusion in this review (). Most of the articles were randomized controlled trials. Data collection periods spanned from 2000 to 2023, providing data from humans and animal models.
Discussion
Food-producing animal complexes: holobionts in a polymicrobial environment
Complex respiratory diseases are a significant cause of morbidity and mortality in livestock, in which prevention, prompt diagnosis, and targeted treatments are essential to keep animal health and welfare (Ericsson et al. Citation2016; Bond et al. Citation2017; Oladunni et al. Citation2019; Ericsson et al. Citation2020; Mach et al. Citation2021). For example, the bovine respiratory disease complex (BRDC) is a leading cause of morbidity and economic losses in wealthy countries (Arcangioli et al. Citation2008; Salem et al. Citation2020; Gaudino et al. Citation2023), especially for young feedlot calves, ranging from 30% in Belgium (van Leenen et al. Citation2020), to 49% in Switzerland, and up to 90% in the U.S.A (Hilton Citation2014). Additionally, sheep respiratory disease affects many animals (Lacasta Citation2019) and causes significant losses, such as carcass condemnations, treatments, and decreased production (Lacasta Citation2019). Moreover, the prevalence of the porcine respiratory disease complex (PRDC) in finishing pigs continues to grow (Qin et al. Citation2018), with a morbidity rate ranging from 10% in Denmark (Hansen et al. Citation2010) to 40% in the U.S.A (Harms et al. Citation2002). As for common livestock animals, the respiratory disease complex in commercial birds remains widespread in countries (Umar et al. Citation2016; Guinat et al. Citation2018; Belkasmi et al. Citation2020; Filaire et al. Citation2022), causing subclinical infections, mild respiratory symptoms, and high production losses in birds raised for meat or eggs (Awad et al. Citation2014; Guabiraba and Schouler Citation2015; Patel et al. Citation2018; Samy and Naguib Citation2018).
These food-producing animal complexes depend on more than one pathogen (Vayssier-Taussat et al. Citation2015). Synergistic interactions between multiple pathogens often occur (Mach and Clark Citation2017; Mazel-Sanchez et al. Citation2019). A plethora of examples in ruminants and swine illustrate the framework for coinfection between pathogens (Gaudino et al. Citation2023), especially in crowded conditions and breeding programs that are overly focused on enhancing traits related to production instead of robustness and resilience. In this context, multiple viral agents can contribute to the development of BRDC (Holman et al. Citation2015; Alexander et al. Citation2020; Chai et al. Citation2022), including bovine viral diarrhea virus (BVDV), bovine respiratory syncytial virus (BRSV), bovine herpes virus 1 (BHV-1), influenza D virus (IDV) (Oliva et al. Citation2019; Lion et al. Citation2021), bovine coronavirus (BCoV) (Salem et al. Citation2020), parainfluenza three virus (PI3V) (Klima et al. Citation2014; Gaudino et al. Citation2023), and Mycoplasma bovis (Lion et al. Citation2021; Gaudino et al. Citation2023). In France, Mycoplasma bovis is the most frequently isolated etiologic agent in these BRDC outbreaks, spreading early and widely throughout the affected units (60–100% isolation rate and seroconversion) (Arcangioli et al. Citation2008). Mannheimia haemolytica is the primary causative pathogen leading to lung damage in sheep (Gupta et al. Citation2023). Mycoplasma ovipneumoniae, PI3V, and M. haemolytica infection predispose sheep to physical and welfare impairment (Sharp et al. Citation1978; Gupta et al. Citation2023). A synergy between nasal Staphylococcus aureus and pathobionts such as Pasteurella multocida and Klebsiella spp. has also been reported in pigs (Espinosa-Gongora et al. Citation2016). The co-occurrence of the porcine reproductive and respiratory syndrome virus (PRRSV), Haemophilus parasuis, and Mycoplasma hyorhinis in lungs is frequently observed (Jiang et al. Citation2019). The swine influenza virus enhances the morbidity of Streptococcus suis infection (Meng et al. Citation2016). Another case in point is that during outbreaks, the low pathogenic avian influenza viruses (LPAIV) are often coupled with coinfections by Mycoplasma gallisepticum, Mycoplasma synoviae, Ornithobacterium rhinotracheale, avian pathogenic Escherichia coli (APEC) and Staphylococcus aureus, which increases the animal’s mortality rate (Much et al. Citation2002; Umar et al. Citation2016; Filaire et al. Citation2022) and reduce productivity (Umar et al. Citation2016). M. gallisepticum, in turn, changes the whole respiratory bacterial community, leading to tracheal inflammation injury and oxidative stress (Miao et al. Citation2022). Frequently, these polymicrobial infections significantly hinder therapy, prognosis, and overall disease management.
Airway mucins: the first line of defense in the respiratory tract
The respiratory tract resists environmental injury despite continuous exposure to pathogens, particles, and toxic chemicals in inhaled air (Fahy and Dickey Citation2010). Such resistance is highly dependent upon adequate mucus (Fahy and Dickey Citation2010) that shields the lungs from environmental insults through a process known as mucociliary clearance (Song et al. Citation2022). Mucus consists of mostly water (approximately > 97%), mucins, non-mucin proteins, ions, lipids, and immunological factors (Fahy and Dickey Citation2010; Bansil and Turner Citation2018; Nason et al. Citation2021).
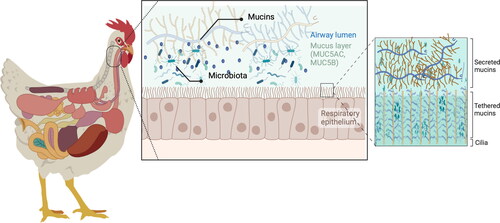
Mucins are the primary structural component of mucus and are made of O-linked glycoproteins (Ridley and Thornton Citation2018) (). There are two classes of mucins: those that remain tethered to cell membranes and those secreted, usually by the goblet cells. The cell-tethered mucins form the basis of a gel-like layer surrounding the cilia (periciliary layer), essential to move mucus out of the airways (Ridley and Thornton Citation2018). In contrast, the secreted mucins constitute the mobile mucus layer (Button et al. Citation2012). The major mucins produced in the airways are the secreted polymeric mucins MUC5AC and MUC5B and the cell-tethered mucins MUC1, MUC4, MUC16, and MUC20 (Thornton et al. Citation2008). In humans, MUC5AC is mainly synthesized in the epithelial surface of goblet cells in the upper airways, whereas MUC5B is primarily secreted from mucous cells in submucosal glands (Thornton et al. Citation2008). Similar conformation is observed in pigs (Ermund et al. Citation2017; Caballero et al. Citation2021) and poultry (Reddy et al. Citation2017). Interestingly, host species differ in genetic makeup and expression of mucins, resulting in a species-specific mucin and glycan repertoire (Pajic et al. Citation2022). The mucus layer can, therefore, be regarded as a significant host range determinant (Wallace et al. Citation2021). However, in all species, mucins have a central protein core rich in Ser-, Pro-, or Thr-repetitive and non-repetitive sequences (Ridley and Thornton Citation2018) decorated with many glycan chains. The glycans attached to the protein mucin core are highly O-glycosylated (Ridley and Thornton Citation2018). These O-glycans are primarily built from five monosaccharide components: galactose, N-acetylglucosamine (GlcNAc), N-acetylgalactosamine (GalNAc), fucose, and sialic acid (Hansson Citation2019), which are attached to the protein backbone through an oxygen atom (Varki Citation2007). Each possible glycan form has a potentially unique regulatory capability (Jin et al. Citation2017). The ensemble of glycan forms found in mucins and their arrangements, particularly within glycoproteins, comprise the glycome (Kunej Citation2019; Shipunov and Kupaev Citation2022). The study of the glycome, the so-called glycomics, is metaphorically akin to forestry; each glycoprotein contains glycans (leaves) conjugated to a protein (tree trunk) (Critcher et al. Citation2022). Briefly, glycomics evaluates the structures and function of glycoproteins in a biological system (Kunej Citation2019) and has started to delineate the emerging and promising role of the mucinome, which is the ensemble of glycoproteins whose mucin domains make them functional (Malaker et al. Citation2022). Glycomics has started identifying human and mouse airway mucin glycan patterns (Walther et al. Citation2013 ;Jia et al. Citation2020; Shipunov and Kupaev Citation2022). Nonetheless, information is scant for livestock. For example, only two studies have described the N-glycan patterns, one in pig (Byrd-Leotis et al. Citation2014) and another in chicken (Suzuki et al. Citation2022) lungs, with no information about the O-glycome. The N- and O-glycans differ in structure, attachment sites, and functions.
Figure 2. Schematic diagram of the respiratory mucus layer.
The mucus covers the respiratory epithelia and consists of two layers: the gel layer of secreted mucins and the periciliary layer of cell-tethered mucins. The gel layer contains soluble mucins MUC5AC and MUC5B. These soluble mucins significantly contribute to this layer’s viscosity and gel-like properties. Cilia on the epithelial surface are rich in MUC1, MUC4, MUC16, and MUC20. Host species differ in genetic makeup and mucin expression, resulting in a species-specific mucin and glycan repertoire on cell surface receptors and in the gel layer. This figure has been created with BioRender.com.
Gaining a complete understanding of the mucin glycome’s role in respiratory health is crucial because an essential function of glycans in airway mucins is to prevent respiratory infections (Schnaar Citation2015; Reily et al. Citation2019; Qin and Mahal Citation2021; Brazil and Parkos Citation2022). Mainly, airway mucin glycosylation patterns are partially responsible for mucus’s ability to set protective physical barriers against mechanical and chemical damage from the external environment (Goto et al. Citation2016) and harmful microorganisms (Xia et al. Citation2005). The terminal sialylated glycans, e.g. sialic acids (Sia), are attached to secreted and tethered mucins at the cell surface and typically bind and trap pathogens, preventing them from accessing the respiratory epithelium (Varki Citation2007). Glycans on mucins also possess antimicrobial properties and modulate and maintain immune homeostasis (Schnaar Citation2015; Reily et al. Citation2019; Qin and Mahal Citation2021; Brazil and Parkos Citation2022). They also prevent opportunistic microbes’ virulence (Kavanaugh et al. Citation2014; Wheeler et al. Citation2019), aggregation, and biofilm formation, interfering with pathogen adhesion and cell receptor binding (Wheeler et al. Citation2019).
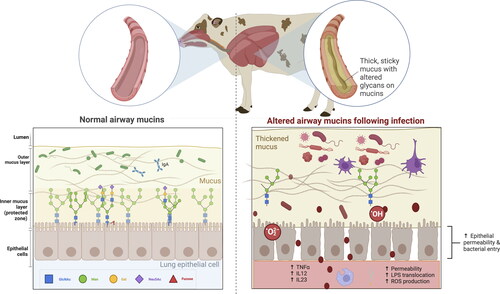
Despite the various defense mechanisms of mucins, some pathogens have developed strategies to exploit or manipulate mucins to enhance their survival or evade host immune responses. For example, Gram-negative and Gram-positive bacteria, including Pseudomonas aeruginosa and Staphylococcus aureus, induce excessive MUC5B production (Roy et al. Citation2014; Fan et al. Citation2022). Excessive mucus production can no longer attenuate microbial virulence and pacify opportunistic pathogens (Schulz et al. Citation2007) (). Bordetella bronchiseptica increases MUC1 production in swine nasal turbinate cells (Park et al. Citation2020). In line with this, viruses can also stimulate MUC5AC overproduction in vitro or in vivo on airway epithelial cells (Thai et al. Citation2008). Likewise, Mycoplasma gallispeticum, one of the most common pathogens in chicken farms, alters the expression of different airway mucin genes, leading to an excessive swelling of the infraorbital sinuses (Miao et al. Citation2022). In extremis, pathogens such as Bordetella pertussis and Pseudomonas aeruginosa can upregulate MUC2 expression in lung goblet cells, inducing mucus overproduction, airway obstruction, and even death (Li et al. Citation1998). Other viruses have evolved surfaces that do not adhere to mucins (Schaefer and Lai Citation2022). In contrast, some pathogens can degrade the mucus layer to penetrate and release toxins that disrupt the epithelial barrier and the porous size or modify mucus pH, influencing its viscosity, ionic strength, and charge. The combination of mucin-degrading proteases, chemotaxis, and flagella allows pathogens to move inside the mucus, adhere to cells, and produce infection (Paone and Cani Citation2020). Several canine mycoplasmas, such as M. canis, M. cynos, and M. molare (May and Brown Citation2009), synthesize sialidase to catalyze the hydrolysis of sialic acid, which aids in the destruction of the extracellular matrix and, therefore, facilitates coinfection. Other viruses, such as influenza viruses, some coronaviruses, and paramyxoviruses, can thwart the mucin barrier by releasing themselves from decoy receptors via activating their associated glycan-receptor-destroying enzymes (Wallace et al. Citation2021).
Figure 3. Barrier function, mucus production, mucociliary clearance, and host defenses in the normal and disturbed airway epithelium.
Panel 1. In a healthy stage, the mobile mucus layer forms a 3-dimensional network in the respiratory tract lumen containing secreted mucins and the residing microbiota. The heavily glycosylated and sialylated mucins (both tethered and secreted) create a barrier against pathogens, which are at risk of being expelled by mucociliary clearance after immobilization in the mucus layer.
Panel 2. Despite mucins’ various defense mechanisms, some pathogens have developed strategies to exploit or manipulate mucins (e.g. increase mucus production or modify glycan profiles) to enhance their survival or to evade host immune responses. The absence or presence of glycotopes in mucins, including sialic acid modifications, are likely to affect the infection potential of viruses. This figure has been created with BioRender.com.
Once the mucus barrier is surpassed, pathogens can recognize and target particular classes of epithelial cell receptors, such as sialylated glycans and glycosaminoglycans, to mediate cellular attachment and entry (Varki Citation2007; Stencel-Baerenwald et al. Citation2014). Several RNA and DNA viruses use Sia to access the host cells as initial anchors (Huang et al. Citation2016). Given their terminal location and wide distribution, Sias are common binding targets for many pathogens (Wasik et al. Citation2016). Binding specificity goes beyond the presence or absence of an appropriate Sia receptor as it depends on the types of Sia, their modifications, and their linkage to the underlying sugar chain (Varki Citation2008). Species-specific differences in the sialoglycan profile probably contribute to mucus being a host range determinant (Wallace et al. Citation2021). The best-known role of sialic acids is in binding influenza and coronaviruses, two of the most critical zoonotic threats, to airway epithelium (Byrd-Leotis et al. Citation2014). Sia linkage type (α2,3 or α2,6) is the hallmark structural element governing the binding affinity of influenza A viruses (IAV; α2,3 binding by avian viruses; α2,6 binding by human, bovine, and swine viruses) (Wallace et al. Citation2021). When studying the Sia structures in the chicken trachea and lung, IAVs bind preferentially to the terminal N-acetylneuraminic acid (Neu5Ac), a type of sialic acid linked to the penultimate galactose through an α2,3 bond, with or without fucosylation and 6-sulfation, but not to an α2,6 bond (Muranaka et al. Citation2011; Suzuki et al. Citation2022). Instead, the influenza D virus in cattle, domestic pigs, goats, sheep, and horses attaches to the 9-O-acetyl form of α2,3 or α2,6-Neu5Ac (Su et al. Citation2017). All influenza viruses tested in pigs interacted with one or more sialylated N-glycans but not O-glycans or glycolipid-derived glycans (Byrd-Leotis et al. Citation2014). Similarly, the 9-O-acetyl form Sia can be used as a specific receptor for bovine coronaviruses (Vlasak et al. Citation1988). Bovine and porcine rotaviruses strains, double-stranded RNA (dsRNA) viruses in the reovirus family use the Sia N-glycolylneuraminic acid (Neu5Gc) instead (Yu et al. Citation2012).
Pathogenic bacteria also use Sias for their benefit as a nutrient and to coat themselves with the molecule, avoiding phagocytosis and the lytic action of the complement cascade. Other pathogens often carry glycan structures on their surface, such as sialic acid-specific glycans, to aid in host cell attachment and to evade host immunity (Severi et al. Citation2007; Chang et al. Citation2014; Schnaar Citation2015; Macauley et al. Citation2015; de Jong et al. Citation2022). For example, virulent strains of Haemophilus parasuis present the sialyltransferase-encoding gene lsgB gene, which is involved in the terminal sialylation of the lipooligosaccharides (Martínez-Moliner et al. Citation2012). This sialylation is a molecular mimicry mechanism to evade the host immune system. Thus, pathogens partake in molecular mimicry by modifying their surface glycans to resemble those of host cells, making it difficult for the immune system to recognize and target them.
In summary, pathogens exploit their own and host mucin glycans to establish infection and survival. The presence of glycotopes on tethered or secreted mucins can have complex and context-dependent effects on health and pathogen interactions. Glycotopes can influence pathogen attachment and entry, immune responses, and the overall susceptibility of animals to infection. Understanding these interactions is crucial for developing strategies to prevent infectious diseases and may have implications for prevention development and therapeutics.
Airway mucins: sweet and well-coated partners for respiratory microbiota
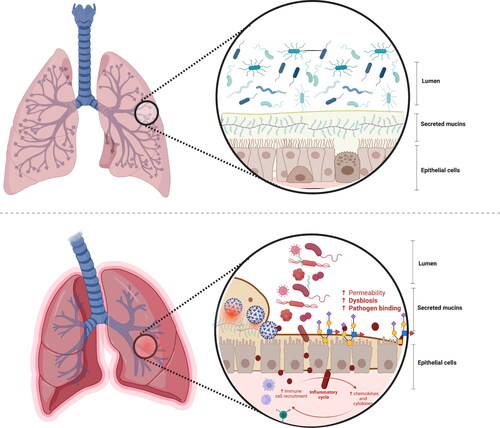
Being both an environmental niche and a food source, respiratory mucins are essential drivers of the airway microbiota composition, diversity, stability, and functionality (Varki Citation2007) (). This is especially true for the airway microbiota, which primarily extracts nutrients from the respiratory mucins because nutrients are scarce.
Figure 4. The microbiome-mucin axis in the respiratory tract: mucin damage matters.
Upper panel: The mutualistic relationship between the airway mucins and the microbiota and derived metabolites under eubiosis. On the one hand, the microbiota and their metabolism induce the synthesis of large gel-forming mucins, including encapsulation, glycosylation, changes in fucosylation and sialidation patterns, and thickness. On the other hand, the mucin layer serves as an environmental niche and a food source for the microbiota. The high diversity of gut mucins impacts the gut microbiota composition, diversity, and stability but also influences immune homeostasis.
Bottom panel: The mutualistic relationship between the mucin glycome and the microbiota under a disrupted airway ecosystem and environment. The deterioration of the respiratory mucosal barrier enables virus binding to the cells and the translocation of bacteria and lipopolysaccharides outside the respiratory tract, triggering immune and inflammatory responses, often resulting in increased permeability and, eventually, endotoxemia. Changes in the respiratory barrier integrity involve changes in the abundance, expression, and glycosylation of mucins, and thus immune dysregulation, dysbiosis, and risk of disease onset.
This figure has been created with BioRender.com.
Although the respiratory tract microbiome is less diverse than the gut (Mach et al. Citation2021), the upper respiratory tract contains a significant microbial biomass (Zeineldin et al. Citation2019). The colonization of the airways occurs shortly after birth in animals, and maturation of the respiratory microbiome can occur within just weeks in ruminants, pigs, and birds (Mach et al. Citation2021). The airway microbiota composition is mediated mainly by microbial immigration, microbial elimination, and the proliferation rate of bacteria (Mach et al. Citation2021). The airway microbiota is essential for proper lung development (Gollwitzer et al. Citation2014), immune responses, and overall health (Scher et al. Citation2016; Zeineldin et al. Citation2019).
The first insights into the respiratory mucin-microbiome interplay in livestock are reported in . Up to date, only five studies have studied the modifications of airway mucins and the microbiota in livestock. For instance, microbiota modification due to different levels of ammonia concentrations in growing pigs impacted the thickness and viscosity of the mucus layer and increased the colonization of harmful bacteria (Wang et al. Citation2019). Furthermore, enriching the upper respiratory tract with the probiotic Bacillus amyloliquefaciens in chicken increased the count of goblet cells and the expression of the MUC2 gene in the tracheal epithelium and the overall respiratory mucosal barrier function (Luan et al. Citation2019). According to these results, nasal probiotic sprays containing Bacillus and Lactobacillus species limited the microscopic damages in the respiratory tract caused by avian influenza virus H9N2, improved the health status of the cilia and the number of goblet cells and epithelial cells in the trachea (Rasaei et al. Citation2023). Further, the interplay between the respiratory microbiota and the mucosal barrier function against pathogens has been recently demonstrated by Miao et al. (Citation2022). These authors showed that transplantation of respiratory microbiota from the Mycoplasma gallisepticum-infected chicks induced inflammatory injury, oxidative stress, mucosal barrier damage, and reduction of microbiota diversity in the receptor animals. Specifically, the microbiota transplantation induced down-regulating tight junction-related and mucin genes, which could be the possible causes of dysregulated immune responses (Ericsson et al. Citation2016; Blakebrough-Hall et al. Citation2020) as well as a decrease in microbiota diversity.
Table 1. First insights into the respiratory mucin-microbiome interplay in livestock.
The tracheal tissue samples
To better understand the mucin-microbiome synergy, one must first investigate how and which mucin glycans shape and drive respiratory microbial composition and function (Belzer et al. Citation2017). Microorganisms are required to synthesize large gel-forming mucins, encapsulation, glycosylation, changes in fucosylation, sialidation patterns, and thickness (Arike et al. Citation2017). Looking deeper at the sheer amount of data we have gathered from the mucin-microbiome interplay in the gut, we know that the microbial ecosystem influences the overall gut barrier function, including mucus layer composition and thickness, intestinal epithelial tight junction protein structure, antimicrobial peptide secretion, and goblet cells abundance at the transcriptional and epigenetic levels. The short-chain fatty acids (SCFAs) butyrate and propionate epigenetically regulate MUC2 gene expression in the human goblet cell-like LS174T cells (Burger-van Paassen et al. Citation2009), which helps maintain intestinal barrier function. In line with these findings, an elegant study by Bergstrom and Xia (Bergstrom and Xia Citation2022) showed that the SCFAs resulting from O-glycan fermentation regulated intestinal mucin barrier function. Bioactive SCFA administration (primarily butyrate) also promotes MUC2 and MUC5AC gene expression and increases epithelial cell integrity after damage (Giromini et al. Citation2022). A study in piglets concluded that gastric infusions of SCFAs maintained intestinal barrier function by increasing the expression of intestinal epithelial tight junction proteins occludin and claudin-1 genes and decreasing the gene and protein abundances of IL-1 in the colon, coupled with reduced intestinal epithelial cell apoptosis (Diao et al. Citation2019). In this regard, the microbiota, the microbial metabolites, the mucins, the epithelium cells, and tight junctions are likely interdependent, so the loss of one diminishes the other (Capaldo et al. Citation2017). Taking advantage of human intestinal enteroids, Pearce et al. (Citation2020) demonstrated that the infusion of microbial-derived SCFA affected the overall gut barrier function, including increased expression of mucin genes, goblet cell markers, common mucus constituents, and antimicrobial peptides (Pearce et al. Citation2020). Butyrate administration in Caco-2 cell monolayers restored occludin and F-actin delocalization and enhanced tight junction protein expression (Peng et al. Citation2009). Besides SCFAs, bacteria and microbial products such as LPS, flagellin, and lipoteichoic acid, among others, can also modify mucin composition and structure. Accordingly, an in vitro study in human mucin-secreting goblet cell line HT29-MTX revealed that bacterial LPS increased mucin MUC5AC, MUC5B, and cytokine mRNA expression (Smirnova et al. Citation2003). Interestingly, co-culturing experiments of Akkermansia muciniphila with non-mucus-degrading butyrate-producing bacteria (Belzer et al. Citation2017) indirectly stimulated intestinal butyrate levels near the intestinal epithelial cells with potential health benefits to the host. Confirming these findings, exposing human enteroids to cecal contents obtained from mice treated with A. muciniphila stimulated the MUC2 protein and MUC2 mRNA expression, along with mucus thickness, compared to the control (Kim et al. Citation2021).
The mucin-microbiome data from gut studies undoubtedly opens possibilities to explore terra incognita in the respiratory health and disease contexts for animals of veterinary interest.
The mucin-microbiome interplay is malleable
The problems mucus and mucins can cause are evident during upper respiratory tract colds (Hansson Citation2019).
Persistent mucus accumulation and airway blockage pervasively impair microbial clearance, enhance the transition from the microbiome to the pathobiome (Fahy and Dickey Citation2010; Ridley and Thornton Citation2018), and increase inflammation (Rose and Voynow Citation2006; Meldrum and Chotirmall Citation2021). The conversion from healthy to pathologic mucus occurs through multiple mechanisms (Fahy and Dickey Citation2010), including abnormal secretion of salt and water, increased submucosal gland mucus secretion (Hoegger et al. Citation2014), mucus infiltration with inflammatory cells, and heightened broncho-vascular permeability with respiratory distress (Fahy and Dickey Citation2010) ().
Beyond mucin expression alterations, glycan glycosylation patterns have also been linked to respiratory physiopathology (Hoffman et al. Citation2020; Shipunov and Kupaev Citation2022). During inflammation, mucin glycans exhibit reduced chain length, sulfation, fucosylation, and increased sialylation (Schulz et al. Citation2007). It is important to note that seemingly minor differences in glycan structures may result in significant pathophysiological outcomes. As an illustration, a shift in the nine-carbon backbone monosaccharides of the sialic acid might enhance the virus binding and infection of cells, facilitating increased colonization and the development of lung disease (Carnoy et al. Citation1994). The specific roles of mucins and their glycome during respiratory infections can vary depending on the pathogen, tissue, and host factors involved (Hoffman et al. Citation2020).
Given the intertwined relationship between the mucin and microbiota and the considerable malleability of the microbiota relative to host genomes, the possibility of influencing respiratory tract health via microbiota manipulation seems possible (Hoffman et al. Citation2020). Today, controlling pathogens’ access to glycans on mucins via microbiome modifications has proven to be a promising method to prevent infection (Hoffman et al. Citation2020). An elegant study by Pereira et al. (Pereira et al. Citation2020) demonstrated that the administration of a synthetic bacterial consortium decreases the availability of sialic acid from mucins by cross-feeding microbes, which consequently protects against an infection caused by microbial pathogens that use these sugar groups as binding sites, e.g. viruses such as influenza virus, reovirus, adenovirus, and rotavirus (Stencel-Baerenwald et al. Citation2014). Therefore, synthetic microbial communities expressing sialidase activity might improve mucosal health and prevent complex respiratory diseases (Almagro-Moreno and Boyd Citation2009). Complementary to this approach, nebulizing heparan sulfate-consuming commensal bacteria consistently contained SARS-CoV-2 attachment in higher-risk individuals (Clausen et al. Citation2020). At the same time, nebulized fucose has been shown to reduce bacterial adhesion and improve lung function in animal models of respiratory infections (Hauber et al. Citation2008). Determining the ideal microbial composition for optimal respiratory health and immune function in livestock is still a topic of ongoing research (Holman et al. Citation2015; Amat et al. Citation2017; Bond et al. Citation2017; Klima et al. Citation2019; Zhang et al. Citation2019; McMullen et al. Citation2020; Guo et al. Citation2020; Alexander et al. Citation2020; Mahmmod et al. Citation2020; Pirolo et al. Citation2021; Chai et al. Citation2022). The nature and the mechanisms behind respiratory microbiome and its interactions with the mucins and glycome profiles in livestock are not understood. Yet, Chen and colleagues have demonstrated in chicks that mulberry leaf polysaccharide (a type of glycan) has the potential to modify mucin structure and activate respiratory mucosal immunity against Newcastle disease virus, possibly via modifications in the airway microbiota (Chen et al. Citation2021). It seems likely that there is a substantial amount of untapped potential concerning the airway microbiome-mucin interplay in the respiratory tract of livestock and how external factors, such as the administration of nebulized synthetic bacterial consortium or the use of food containing synthetic and natural dietary glycans designed to target microbial activity at the mucosa. In the gut, such approaches have shed light on how alterations to the biochemistry of mucins and mucus impact their protective capacity and restore healthy mucosal function (Belzer et al. Citation2017; Ottman et al. Citation2017; Belzer Citation2022). Several examples of dietary glycans already show their potential to resolve mucus-layer defects associated with disorders such as type 2 diabetes, obesity, and gastrointestinal pathologies (e.g. Crohn’s disease, ulcerative colitis, and colorectal cancer) (reviewed by (Clark and Mach Citation2023)).
Lastly, a new avenue is understanding if gut microbiota-derived metabolites could be critical molecular mediators of the airway microbiota-mucin interplay and could affect the onset of respiratory diseases. It has been recently suggested that respiratory comorbidities might be partly regulated by the bidirectional gut-lung intercommunication (Dang and Marsland Citation2019), the so-called gut-lung axis (Enaud et al. Citation2020). This bidirectional communication likely occurs via the translocation of gut microbes and metabolites and local and systemic immune responses (Budden et al. Citation2017). Research is now deciphering how the gut microbiota and its metabolites modulate the onset of respiratory infections and whether the airway microbiota, in turn, influences immune cells to adjust inflammatory responses at distal sites such as the gut (Wypych et al. Citation2019). For instance, in a study developed in chickens, Lactobacillus salivarius intake alleviated lung inflammation injury caused by Mycoplasma gallisepticum infection and increased host defense against Escherichia coli by improved gut microbiota composition (Wang et al. Citation2021). More examples of livestock are reported elsewhere (Mach et al. Citation2021).
Non-coding RNAs: controlling mucin production and glycome patterns
Understanding the genetic and genomic regulation of mucin production and glycosylation patterns offers another way to modulate mucin levels, affecting specific signaling pathways or transcription factors involved in their synthesis and glycosylation, among others. However, transmembrane and secreted mucins have a complex glycosylated nature and extreme size (Gum Citation1992), and they are products of an orchestrated collection of enzymes working in a coordinated manner (Thu and Mahal Citation2020).
As conserved regulatory agents, ncRNAs have gained attention as crucial mucin and glycome regulators at the transcriptional, posttranscriptional, and translational levels. The ncRNAs are RNA molecules that do not encode proteins but have regulatory or structural functions within cells (Kosinska-Selbi et al. Citation2020). ncRNAs are mainly divided into small ncRNAs and long ncRNAs. MicroRNAs, circular RNAs, and their precursor pri-miRNAs are small ncRNAs (sncRNAs) ranging from ∼22 bp in their mature form to ∼70 bp in their premature stem-loop form. LncRNAs are long ncRNAs transcribed by RNA polymerase II (RNA Pol II) and are longer than 200 nucleotides. Like miRNAs, lncRNAs have emerged as new regulators of gene expression and have become a focal point of biomedical and veterinary research (Do et al. Citation2021).
MiRNAs regulate gene expression at the post-transcriptional level by binding (usually with imperfect complementarity) to the 3-UTR of a target mRNA, resulting in translation degradation or inhibition. A single miRNA can have hundreds of target genes, and multiple miRNAs can converge on a single mRNA. Unlike transcriptional regulators, the role of miRNAs is not to turn a gene on or off but to regulate gene expression (Thu and Mahal Citation2020). On the other hand, the lncRNAs regulate the genomic output at many levels, from transcription to translation (Malmuthuge and Guan Citation2021). The effect of lncRNAs is mainly achieved by interfering with the expression of downstream genes, supplementing or interfering with the mRNA splicing process, and regulating protein activity (Wang et al. Citation2021). An individual transcriptome contains more lncRNAs than mRNA molecules (Foissac et al. Citation2019). For livestock, the most significant number of identified lncRNA transcripts is available for pigs and cattle (Foissac et al. Citation2019). Poultry is represented by less than half of the records. Genomic annotation of lncRNAs showed that most are assigned to introns (pig, poultry) or intergenic (cattle) (Kosinska-Selbi et al. Citation2020). The number of detected miRNAs in farm animal species is lower than lncRNs, from 1,064 in cattle to 267 in goats and 406 in pigs (miRbase release 22; http://www.mirbase.org/). However, miRNAs have received more research attention and scrutiny than lncRNAs in livestock and companion animals (Pacholewska et al. Citation2016; Mach et al. Citation2016).
Given the emerging importance of ncRNAs in health and disease and their potential to modify genetic expression, it is clear that more attention should be paid to ncRNA and mucin interactions in the context of respiratory infections. To this point, data on ncRNAs’ roles in mucin regulation has been mainly generated by well-controlled murine models and humans. In humans, mucin expression during infection is regulated by a panoply of miRNAs, namely miR-34b/c (Li et al. Citation2021), miR-146a (Zhong et al. Citation2011), miR-378 (Skrzypek et al. Citation2013), and miR-141 (Siddiqui et al. Citation2021). MiRNAs can also control the expression of glycosyltransferases and modify the glycome on mucins (Agrawal et al. Citation2014; Thu and Mahal Citation2020). Jame-Chenarboo et al. (Citation2022) established that miRNAs are substantial regulators of cell glycosylation. Therefore, changes in mucin glycosylation patterns, such as increased sialylation, will likely alter mucin’s binding properties with pathogens and their protective functions.
The mechanisms and results based on laboratory animals have yet to be validated in animals of veterinary interest due to the limitations in obtaining samples and the complexity of production systems, hosts, biotic and abiotic stressors, and respiratory infectious etiology. Nevertheless, recent studies indicate that miRNAs tune host mucins to face respiratory pathogen infection in farm animals (). In experimentally infected pigs with the Aujeszky’s disease virus, miR-206 was upregulated and likely interacted with the MUC1 gene, whose primary function is to act as a physical barrier by trapping pathogens (Timoneda et al. Citation2014). Similarly, a set of miRNAs responded to swine Influenza A virus subtype H1N2 infection (e.g. miR-15a, miR-21, miR-146, miR-206, miR-223, and miR-451) by inducing the antimicrobial protein MUC1 (Skovgaard et al. Citation2013). Mucin-type O-glycan biosynthesis pathways were modified at the molecular level via miRNA action following PRRSV infection in pigs. For instance, PRRSV infection upregulated ssc-miR-27b-3p and ssc-miR-23a-3p and induced the expression of mucin type-O pathway (Fleming and Miller Citation2019). In a possible attempt to bolster mucosal immunity, the avian leukosis virus subgroup J also caused changes in the mucin-type O-Glycan biosynthesis in lungs (Zhang et al. Citation2019). Collectively, these studies reveal a role for miRNAs as potent host-derived regulators of respiratory pathogen infection that broadly affect how the host prevents mucosal infections.
Table 2. Functional implications of mucin regulation by ncRNAs in livestock infected with viruses with respiratory tropism.
Much work is needed to explain the influence of ncRNAs on mucin and pathogen virulence and infectivity.
An extra difficulty lies in the fact that the presence and activity of microbiota might also regulate the host ncRNAs (Oliveira et al. Citation2023; Aggeletopoulou et al. Citation2023). Indeed, the microbiomes and microbial metabolites such as secondary bile acids and SCFAs have been shown to regulate the expression of miRNAs (Pant et al. Citation2017) and lncRNAs in the intestine epithelial cells (Dempsey et al. Citation2018), macrophages (Gao et al. Citation2020), and other metabolic organs (Dempsey et al. Citation2018) in adult mice or in vitro (Chowdhury et al. Citation2017) studies, suggesting that the gut microbiome regulates the expression of both coding RNAs and ncRNAs regionally and systemically (Sommer et al. Citation2015). For instance, a study investigating early rumen development in neonatal dairy calves revealed that nearly 46% of miRNAs expressed in the rumen are responsive to SCFA (Malmuthuge et al. Citation2019). Another experiment on rumen microbiota (Ricci et al. Citation2022) suggested that the microbiota influences the host’s miRNA expression patterns and that the host potentially helps shape the gut bacterial profile by producing specific miRNAs. The airway microbiota and ncRNA relationship mechanisms are not yet defined, and no knowledge exists about the ncRNA regulatory mechanisms behind mucin-microbiome interactions in the respiratory tract. Still, these emerging nuances in the gut illustrate that the microbiome might interact with the host genome directly (direct modulation of transcriptome) and indirectly via the expression of ncRNA (Malmuthuge and Guan Citation2021).
Directions for further research
The few studies that satisfied our inclusion criteria have emphasized the requirement for more cohort studies designed to identify respiratory microbiome-mucin modifications and their genomic control to understand better the interplay between pathogens, respiratory health, and the use of interventions such as pre-and probiotics.
New avenues might include the use of cutting-edge tools to describe how and which mucin glycans shape and drive respiratory microbial composition and function under eubiosis in different farm species and dissect the glycan-mediated interactions within the microbiota ecosystem. Integrating insights from diverse fields, such as metagenomics, metatranscriptomics, and glycomics, will foster a comprehensive understanding of glycome-microbiota dynamics. Coupled with this, longitudinal studies analyzing pre- and post-infection modifications of mucin glycan and microbiota profiles in the respiratory tract will enable the identification of the evolving nature of this relationship and discover molecular features related to resilience or recovery within the same subject. Pathogen and species-specific studies, encompassing bacteria and viruses combined, are obliged, as some pathogens likely have more specific effects on the microbiota and mucins than others.
Shifting the focus toward functional characterization of glycan-microbiota interactions, studies designed to explain the influence of non-coding RNAs on holobiont response (at the transcriptional and epigenetic levels) under infection and the mechanisms through which host glycan expression influences microbial composition and pathogen protection are needed. Lastly, compelling studies are also required to evaluate the therapeutic potential of modulating the glycome and the microbiota composition for respiratory health benefits. Experiments designed to decipher the role of nebulized synthetic bacterial consortiums or food containing synthetic and natural dietary glycans on the microbial activity at the mucosa and the holobiont response might have therapeutic implications. By unraveling these aspects, we will comprehensively overview the exciting developments and future directions in mucin-microbiota studies.
Concluding remarks
In livestock pathology, there is a growing awareness that respiratory infectious agents frequently do not operate alone, and their virulence and pathogenicity are likely affected by their interaction with the microbiome-mucin interplay. Therefore, microbiome-mucin interactions are a significant player in the health of the respiratory tract. As such, they contain signals that can be used to identify airway pathophysiology. There are fundamental knowledge gaps for species of veterinary interest. Furthermore, ncRNAs may play a vital role in the microbiota-mucin crosstalk via modifying the holobiont. The next phase in the livestock research area should focus on understanding whether particular microbiome-mucin glycome structures confer resistance and resilience to respiratory pathogen infection and which are the critical factors controlling the mucin-microbiome interconnection, including the role of ncRNAs. It is now time to harness the forecasting power of the microbiome-mucin synergies.
Disclosure statement
The author declares no competing interests.
Additional information
Funding
References
- Aggeletopoulou I, Mouzaki A, Thomopoulos K, Triantos C. 2023. miRNA molecules—late breaking treatment for inflammatory bowel diseases? Int J Mol Sci. 24(3):2233. https://doi.org/10.3390/ijms24032233.
- Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, Batista BS, Eng WS, Hsu K-L, Liang Y, et al. 2014. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc Natl Acad Sci USA. 111(11):4338–4343. https://doi.org/10.1073/pnas.1321524111.
- Alexander TW, Timsit E, Amat S. 2020. The role of the bovine respiratory bacterial microbiota in health and disease. Anim Health Res Rev. 21(2):168–171. https://doi.org/10.1017/S1466252320000316.
- Almagro-Moreno S, Boyd EF. 2009. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 9(1):118. https://doi.org/10.1186/1471-2148-9-118.
- Amat S, Subramanian S, Timsit E, Alexander TW. 2017. Probiotic bacteria inhibit the bovine respiratory pathogen Mannheimia haemolytica serotype 1 in vitro. Lett Appl Microbiol. 64(5):343–349. https://doi.org/10.1111/lam.12723.
- Arcangioli M-A, Duet A, Meyer G, Dernburg A, Bézille P, Poumarat F, Le Grand D. 2008. The role of Mycoplasma bovis in bovine respiratory disease outbreaks in veal calf feedlots. Vet J. 177(1):89–93. https://doi.org/10.1016/j.tvjl.2007.03.008.
- Arike L, Holmén-Larsson J, Hansson GC. 2017. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology. 27(4):318–328. https://doi.org/10.1093/glycob/cww134.
- Atanasova KR, Reznikov LR. 2019. Strategies for measuring airway mucus and mucins. Respir Res. 20(1):261. https://doi.org/10.1186/s12931-019-1239-z.
- Awad F, Chhabra R, Baylis M, Ganapathy K. 2014. An overview of infectious bronchitis virus in chickens. Worlds Poult Sci J. 70(2):375–384. https://doi.org/10.1017/S0043933914000385.
- Bansil R, Turner BS. 2018. The biology of mucus: composition, synthesis and organization. Adv Drug Deliv Rev. 124:3–15. https://doi.org/10.1016/j.addr.2017.09.023.
- Belkasmi SFZ, Fellahi S, Touzani CD, Faraji FZ, Maaroufi I, Delverdier M, Guérin J-L, Fihri OF, El Houadfi M, Ducatez MF, et al. 2020. Co-infections of chickens with avian influenza virus H9N2 and Moroccan Italy 02 infectious bronchitis virus: effect on pathogenesis and protection conferred by different vaccination programmes. Avian Pathol. 49(1):21–28. https://doi.org/10.1080/03079457.2019.1656328.
- Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, de Vos WM. 2017. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio. 8(5):e00770-17. https://doi.org/10.1128/mBio.00770-17.
- Belzer C. 2022. Nutritional strategies for mucosal health: the interplay between microbes and mucin glycans. Trends Microbiol. 30(1):13–21. https://doi.org/10.1016/j.tim.2021.06.003.
- Bergstrom K, Fu J, Johansson MEV, Liu X, Gao N, Wu Q, Song J, McDaniel JM, McGee S, Chen W, et al. 2017. Core 1– and 3–derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 10(1):91–103. https://doi.org/10.1038/mi.2016.45.
- Bergstrom K, Xia L. 2022. The barrier and beyond: roles of intestinal mucus and mucin-type O-glycosylation in resistance and tolerance defense strategies guiding host-microbe symbiosis. Gut Microbes. 14(1):2052699. https://doi.org/10.1080/19490976.2022.2052699.
- Blakebrough-Hall C, McMeniman JP, González LA. 2020. An evaluation of the economic effects of bovine respiratory disease on animal performance, carcass traits, and economic outcomes in feedlot cattle defined using four BRD diagnosis methods. J Anim Sci. 98(2):skaa005. https://doi.org/10.1093/jas/skaa005.
- Bond SL, Timsit E, Workentine M, Alexander T, Léguillette R. 2017. Upper and lower respiratory tract microbiota in horses: bacterial communities associated with health and mild asthma (inflammatory airway disease) and effects of dexamethasone. BMC Microbiol. 17(1):184. https://doi.org/10.1186/s12866-017-1092-5.
- Bond SL, Workentine M, Hundt J, Gilkerson JR, Léguillette R, UCVM Class of 2019. 2020. Effects of nebulized dexamethasone on the respiratory microbiota and mycobiota and relative equine herpesvirus-1, 2, 4, 5 in an equine model of asthma. J Vet Intern Med. 34(1):307–321. https://doi.org/10.1111/jvim.15671.
- Brazil JC, Parkos CA. 2022. Finding the sweet spot: glycosylation mediated regulation of intestinal inflammation. Mucosal Immunol. 15(2):211–222. https://doi.org/10.1038/s41385-021-00466-8.
- Brogaard L, Larsen LE, Heegaard PMH, Anthon C, Gorodkin J, Dürrwald R, Skovgaard K. 2018. IFN-λ and microRNAs are important modulators of the pulmonary innate immune response against influenza A (H1N2) infection in pigs. PLoS One. 13(4):e0194765. https://doi.org/10.1371/journal.pone.0194765.
- Budden KF, Gellatly SL, Wood DLA, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. 2017. Emerging pathogenic links between microbiota and the gut–lung axis. Nat Rev Microbiol. 15(1):55–63. https://doi.org/10.1038/nrmicro.2016.142.
- Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. 2009. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 420(2):211–219. https://doi.org/10.1042/BJ20082222.
- Button B, Cai L-H, Ehre C, Kesimer M, Hill DB, Sheehan JK, Boucher RC, Rubinstein M. 2012. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 337(6097):937–941. https://doi.org/10.1126/science.1223012.
- Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, et al. 2014. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci USA. 111(22):E2241–50. https://doi.org/10.1073/pnas.1323162111.
- Caballero I, Ringot-Destrez B, Si-Tahar M, Barbry P, Guillon A, Lantier I, Berri M, Chevaleyre C, Fleurot I, Barc C, et al. 2021. Evidence of early increased sialylation of airway mucins and defective mucociliary clearance in CFTR-deficient piglets. J Cyst Fibros. 20(1):173–182. https://doi.org/10.1016/j.jcf.2020.09.009.
- Capaldo CT, Powell DN, Kalman D. 2017. Layered defense: how mucus and tight junctions seal the intestinal barrier. J Mol Med (Berl). 95(9):927–934. https://doi.org/10.1007/s00109-017-1557-x.
- Carnoy C, Scharfman A, Van Brussel E, Lamblin G, Ramphal R, Roussel P. 1994. Pseudomonas aeruginosa outer membrane adhesins for human respiratory mucus glycoproteins. Infect Immun. 62(5):1896–1900. https://doi.org/10.1128/iai.62.5.1896-1900.1994.
- Chai J, Capik SF, Kegley B, Richeson JT, Powell JG, Zhao J. 2022. Bovine respiratory microbiota of feedlot cattle and its association with disease. Vet Res. 53(1):4. https://doi.org/10.1186/s13567-021-01020-x.
- Chang Y-C, Olson J, Beasley FC, Tung C, Zhang J, Crocker PR, Varki A, Nizet V. 2014. Group B streptococcus engages an inhibitory siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog. 10(1):e1003846. https://doi.org/10.1371/journal.ppat.1003846.
- Chatterjee M, van Putten JPM, Strijbis K. 2020. Defensive properties of mucin glycoproteins during respiratory infections—relevance for SARS-CoV-2. mBio. 11(6):e02374-20. https://doi.org/10.1128/mBio.02374-20.
- Chen X, Yang H, Jia J, Chen Y, Wang J, Chen H, Jiang C. 2021. Mulberry leaf polysaccharide supplementation contributes to enhancing the respiratory mucosal barrier immune response in Newcastle disease virus–vaccinated chicks. Poult Sci. 100(2):592–602. https://doi.org/10.1016/j.psj.2020.11.039.
- Chowdhury IH, Narra HP, Sahni A, Khanipov K, Schroeder CLC, Patel J, Fofanov Y, Sahni SK. 2017. Expression profiling of long noncoding RNA splice variants in human microvascular endothelial cells: lipopolysaccharide effects in vitro. Mediators Inflamm. 2017:3427461–3427418. https://doi.org/10.1155/2017/3427461.
- Clark A, Mach N. 2023. The gut mucin-microbiota interactions: a missing key to optimizing endurance performance. Front Physiol. 14:1284423. https://doi.org/10.3389/fphys.2023.1284423.
- Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, et al. 2020. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 183(4):1043–1057.e15. https://doi.org/10.1016/j.cell.2020.09.033.
- Correa-Fiz F, dos Santos JMG, Illas F, Aragon V. 2019. Antimicrobial removal on piglets promotes health and higher bacterial diversity in the nasal microbiota. Sci Rep. 9(1):6545. https://doi.org/10.1038/s41598-019-43022-y.
- Correa-Fiz F, Fraile L, Aragon V. 2016. Piglet nasal microbiota at weaning may influence the development of Glässer’s disease during the rearing period. BMC Genomics. 17(1):404. https://doi.org/10.1186/s12864-016-2700-8.
- Critcher M, Hassan AA, Huang ML. 2022. Seeing the forest through the trees: characterizing the glycoproteome. Trends Biochem Sci. 47(6):492–505. https://doi.org/10.1016/j.tibs.2022.02.007.
- Dang AT, Marsland BJ. 2019. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 12(4):843–850. https://doi.org/10.1038/s41385-019-0160-6.
- de Jong H, Wösten MMSM, Wennekes T. 2022. Sweet impersonators: molecular mimicry of host glycans by bacteria. Glycobiology. 32(1):11–22. https://doi.org/10.1093/glycob/cwab104.
- Dempsey J, Zhang A, Cui JY. 2018. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genomics. 19(1):834. https://doi.org/10.1186/s12864-018-5235-3.
- Diao H, Jiao AR, Yu B, Mao XB, Chen DW. 2019. Gastric infusion of short-chain fatty acids can improve intestinal barrier function in weaned piglets. Genes Nutr. 14(1):4. https://doi.org/10.1186/s12263-019-0626-x.
- Do DN, Dudemaine P-L, Mathur M, Suravajhala P, Zhao X, Ibeagha-Awemu EM. 2021. miRNA regulatory functions in farm animal diseases, and biomarker potentials for effective therapies. Int J Mol Sci. 22(6):3080. https://doi.org/10.3390/ijms22063080.
- Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, Delhaes L. 2020. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 10:9. https://doi.org/10.3389/fcimb.2020.00009.
- Ericsson AC, Personett AR, Grobman ME, Rindt H, Reinero CR. 2016. Composition and predicted metabolic capacity of upper and lower airway microbiota of healthy dogs in relation to the fecal microbiota. PLoS One. 11(5):e0154646. https://doi.org/10.1371/journal.pone.0154646.
- Ericsson AC, Personett AR, Rindt H, Grobman ME, Reinero CR. 2020. Respiratory dysbiosis and population-wide temporal dynamics in canine chronic bronchitis and non-inflammatory respiratory disease. PLoS One. 15(1):e0228085. https://doi.org/10.1371/journal.pone.0228085.
- Ermund A, Meiss LN, Rodriguez-Pineiro AM, Bähr A, Nilsson HE, Trillo-Muyo S, Ridley C, Thornton DJ, Wine JJ, Hebert H, et al. 2017. The normal trachea is cleaned by MUC5B mucin bundles from the submucosal glands coated with the MUC5AC mucin. Biochem Biophys Res Commun. 492(3):331–337. https://doi.org/10.1016/j.bbrc.2017.08.113.
- Espinosa-Gongora C, Larsen N, Schønning K, Fredholm M, Guardabassi L. 2016. Differential analysis of the nasal microbiome of pig carriers or non-carriers of staphylococcus aureus. PLoS One. 11(8):e0160331. https://doi.org/10.1371/journal.pone.0160331.
- Fahy JV, Dickey BF. 2010. Airway mucus function and dysfunction. N Engl J Med. 363(23):2233–2247. https://doi.org/10.1056/NEJMra0910061.
- Fan L, Lu Y, Wang Y, Zhang X, Wu Y, Sun H, Zhang J. 2022. Respiratory MUC5B disproportion is involved in severe community-acquired pneumonia. BMC Pulm Med. 22(1):90. https://doi.org/10.1186/s12890-022-01870-x.
- Filaire F, Lebre L, Foret-Lucas C, Vergne T, Daniel P, Lelièvre A, de Barros A, Jbenyeni A, Bolon P, Paul M, et al. 2022. Highly pathogenic avian influenza A(H5N8) clade 2.3.4.4b virus in dust samples from poultry farms, France, 2021. Emerg Infect Dis. 28(7):1446–1450. https://doi.org/10.3201/eid2807.212247.
- Fleming DS, Miller LC. 2019. Differentially expressed MiRNAs and tRNA genes affect host homeostasis during highly pathogenic porcine reproductive and respiratory syndrome virus infections in young pigs. Front Genet. 10:691. https://doi.org/10.3389/fgene.2019.00691.
- Foissac S, Djebali S, Munyard K, Vialaneix N, Rau A, Muret K, Esquerré D, Zytnicki M, Derrien T, Bardou P, et al. 2019. Multi-species annotation of transcriptome and chromatin structure in domesticated animals. BMC Biol. 17(1):108. https://doi.org/10.1186/s12915-019-0726-5.
- Gaeta NC, Lima SF, Teixeira AG, Ganda EK, Oikonomou G, Gregory L, Bicalho RC. 2017. Deciphering upper respiratory tract microbiota complexity in healthy calves and calves that develop respiratory disease using shotgun metagenomics. J Dairy Sci. 100(2):1445–1458. https://doi.org/10.3168/jds.2016-11522.
- Gao Y, Zhou J, Qi H, Wei J, Yang Y, Yue J, Liu X, Zhang Y, Yang R. 2020. LncRNA lncLy6C induced by microbiota metabolite butyrate promotes differentiation of Ly6Chigh to Ly6Cint/neg macrophages through lncLy6C/C/EBPβ/Nr4A1 axis. Cell Discov. 6(1):87. https://doi.org/10.1038/s41421-020-00211-8.
- Gaudino M, Lion A, Sagné E, Nagamine B, Oliva J, Terrier O, Errazuriz-Cerda E, Scribe A, Sikht F-Z, Simon E, et al. 2023. The activation of the RIG-I/MDA5 signaling pathway upon influenza D virus infection impairs the pulmonary proinflammatory response triggered by mycoplasma bovis superinfection. J Virol. 97(2):e0142322. https://doi.org/10.1128/jvi.01423-22.
- Gaudino M, Valarcher J-F, Hägglund S, Näslund K, Zohari S, Ducatez MF, Meyer G. 2023. Molecular and genetic characterization of bovine parainfluenza type 3 European field and vaccine strains. Infect Genet Evol. 113:105483. https://doi.org/10.1016/j.meegid.2023.105483.
- Giromini C, Baldi A, Rebucci R, Lanzoni D, Policardi M, Sundaram TS, Purup S. 2022. Role of short chain fatty acids to counteract inflammatory stress and mucus production in human intestinal HT29-MTX-E12 cells. Foods. 11(13):1983. https://doi.org/10.3390/foods11131983.
- Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, Nicod LP, Lloyd CM, Marsland BJ. 2014. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 20(6):642–647. https://doi.org/10.1038/nm.3568.
- Goto Y, Uematsu S, Kiyono H. 2016. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol. 17(11):1244–1251. https://doi.org/10.1038/ni.3587.
- Guabiraba R, Schouler C. 2015. Avian colibacillosis: still many black holes. FEMS Microbiol Lett. 362(15):fnv118. https://doi.org/10.1093/femsle/fnv118.
- Guinat C, Nicolas G, Vergne T, Bronner A, Durand B, Courcoul A, Gilbert M, Guérin J-L, Paul MC. 2018. Spatio-temporal patterns of highly pathogenic avian influenza virus subtype H5N8 spread, France, 2016 to 2017. Eurosurveillance. 23(26):1700791. https://doi.org/10.2807/1560-7917.ES.2018.23.26.1700791.
- Gum JR. 1992. Mucin genes and the proteins they encode: structure, diversity, and regulation. Am J Respir Cell Mol Biol. 7(6):557–564. https://doi.org/10.1165/ajrcmb/7.6.557.
- Guo Y, McMullen C, Timsit E, Hallewell J, Orsel K, van der Meer F, Yan S, Alexander TW. 2020. Genetic relatedness and antimicrobial resistance in respiratory bacteria from beef calves sampled from spring processing to 40 days after feedlot entry. Vet Microbiol. 240:108478. https://doi.org/10.1016/j.vetmic.2019.108478.
- Gupta SK, Parlane N, Bridgeman B, Lynch AT, Dangerfield EM, Timmer MSM, Stocker BL, Wedlock DN. 2023. The trehalose glycolipid C18Brar promotes antibody and T-cell immune responses to Mannheimia haemolytica and Mycoplasma ovipneumoniae whole cell antigens in sheep. PLoS One. 18(1):e0278853. https://doi.org/10.1371/journal.pone.0278853.
- Hansen MS, Pors SE, Jensen HE, Bille-Hansen V, Bisgaard M, Flachs EM, Nielsen OL. 2010. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J Comp Pathol. 143(2–3):120–131. https://doi.org/10.1016/j.jcpa.2010.01.012.
- Hansson GC. 2019. Mucus and mucins in diseases of the intestinal and respiratory tracts. J Intern Med. 285(5):479–490. https://doi.org/10.1111/joim.12910.
- Harms PA, Halbur PG, Sorden SD. 2002. Three cases of porcine respiratory disease complex associated with porcine circovirus type 2 infection. J Swine Health Prod. 10:27–30.
- Hauber H-P, Schulz M, Pforte A, Mack D, Zabel P, Schumacher U. 2008. Inhalation with fucose and galactose for treatment of Pseudomonas aeruginosa in cystic fibrosis patients. Int J Med Sci. 5(6):371–376. https://doi.org/10.7150/ijms.5.371.
- Hilton WM. 2014. BRD in 2014: where have we been, where are we now, and where do we want to go? Anim Health Res Rev. 15(2):120–122. https://doi.org/10.1017/S1466252314000115.
- Hoegger MJ, Fischer AJ, McMenimen JD, Ostedgaard LS, Tucker AJ, Awadalla MA, Moninger TO, Michalski AS, Hoffman EA, Zabner J, et al. 2014. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science. 345(6198):818–822. https://doi.org/10.1126/science.1255825.
- Hoffman CL, Lalsiamthara J, Aballay A. 2020. Host mucin is exploited by Pseudomonas aeruginosa to provide monosaccharides required for a successful infection. mBio. 11(2):e00060-20. https://doi.org/10.1128/mBio.00060-20.
- Holman DB, McAllister TA, Topp E, Wright ADG, Alexander TW. 2015. The nasopharyngeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet Microbiol. 180(1–2):90–95. https://doi.org/10.1016/j.vetmic.2015.07.031.
- Holt HR, Alarcon P, Velasova M, Pfeiffer DU, Wieland B. 2011. BPEX Pig Health Scheme: a useful monitoring system for respiratory disease control in pig farms? BMC Vet Res. 7(1):82. https://doi.org/10.1186/1746-6148-7-82.
- Huang ML, Fisher CJ, Godula K. 2016. Glycomaterials for probing host–pathogen interactions and the immune response. Exp Biol Med (Maywood). 241(10):1042–1053. https://doi.org/10.1177/1535370216647811.
- Jame-Chenarboo F, Ng HH, Macdonald D, Mahal LK. 2022. High-throughput analysis reveals miRNA upregulating α-2,6-sialic acid through direct miRNA-mRNA interactions. ACS Cent Sci. 8(11):1527–1536. https://doi.org/10.1021/acscentsci.2c00748.
- Jia N, Byrd-Leotis L, Matsumoto Y, Gao C, Wein AN, Lobby JL, Kohlmeier JE, Steinhauer DA, Cummings RD. 2020. The human lung glycome reveals novel glycan ligands for influenza A virus. Sci Rep. 10(1):5320. https://doi.org/10.1038/s41598-020-62074-z.
- Jiang N, Liu H, Wang P, Huang J, Han H, Wang Q. 2019. Illumina MiSeq Sequencing investigation of microbiota in bronchoalveolar lavage fluid and cecum of the swine infected with PRRSV. Curr Microbiol. 76(2):222–230. https://doi.org/10.1007/s00284-018-1613-y.
- Jin C, Kenny DT, Skoog EC, Padra M, Adamczyk B, Vitizeva V, Thorell A, Venkatakrishnan V, Lindén SK, Karlsson NG, et al. 2017. Structural diversity of human gastric mucin glycans. Mol Cell Proteomics. 16(5):743–758. https://doi.org/10.1074/mcp.M116.067983.
- Kavanaugh NL, Zhang AQ, Nobile CJ, Johnson AD, Ribbeck K. 2014. Mucins suppress virulence traits of Candida albicans. mBio. 5(6):e01911. https://doi.org/10.1128/mBio.01911-14.
- Kim S, Shin Y-C, Kim T-Y, Kim Y, Lee Y-S, Lee S-H, Kim M-N, O E, Kim KS, Kweon M-N. 2021. Mucin degrader Akkermansia muciniphila accelerates intestinal stem cell-mediated epithelial development. Gut Microbes. 13(1):1–20. https://doi.org/10.1080/19490976.2021.1892441.
- Klima CL, Holman DB, Ralston BJ, Stanford K, Zaheer R, Alexander TW, McAllister TA. 2019. Lower respiratory tract microbiome and resistome of bovine respiratory disease mortalities. Microb Ecol. 78(2):446–456. https://doi.org/10.1007/s00248-019-01361-3.
- Klima CL, Zaheer R, Cook SR, Booker CW, Hendrick S, Alexander TW, McAllister TA. 2014. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol. 52(2):438–448. https://doi.org/10.1128/JCM.02485-13.
- Kosinska-Selbi B, Mielczarek M, Szyda J. 2020. Review: long non-coding RNA in livestock. Animal. 14(10):2003–2013. https://doi.org/10.1017/S1751731120000841.
- Kuiken T, Leighton FA, Fouchier RAM, LeDuc JW, Peiris JSM, Schudel A, Stöhr K, Osterhaus ADME. 2005. Pathogen surveillance in animals. Science. 309(5741):1680–1681. https://doi.org/10.1126/science.1113310.
- Kunej T. 2019. Rise of systems glycobiology and personalized glycomedicine: why and how to integrate glycomics with multiomics science? OMICS. 23(12):615–622. https://doi.org/10.1089/omi.2019.0149.
- Lacasta D. 2019. Preface: special issue on sheep respiratory diseases. Small Ruminant Res. 181:85–86. https://doi.org/10.1016/j.smallrumres.2019.10.011.
- Li JD, Feng W, Gallup M, Kim JH, Gum J, Kim Y, Basbaum C. 1998. Activation of NF-κB via a Src-dependent Ras-MAPK-pp90rsk pathway is required for Pseudomonas aeruginosa-induced mucin overproduction in epithelial cells. Proc Natl Acad Sci USA. 95(10):5718–5723. https://doi.org/10.1073/pnas.95.10.5718.
- Li N, Ma WT, Pang M, Fan QL, Hua JL. 2019. The commensal microbiota and viral infection: a comprehensive review. Front Immunol. 10:1551. https://doi.org/10.3389/fimmu.2019.01551.
- Li W, Du X, Yang Y, Yuan L, Yang M, Qin L, Wang L, Zhou K, Xiang Y, Qu X, et al. 2021. miRNA‐34b/c regulates mucus secretion in RSV‐infected airway epithelial cells by targeting FGFR1. J Cell Mol Med. 25(22):10565–10574. https://doi.org/10.1111/jcmm.16988.
- Lion A, Secula A, Rançon C, Boulesteix O, Pinard A, Deslis A, Hägglund S, Salem E, Cassard H, Näslund K, et al. 2021. Enhanced Pathogenesis Caused by Influenza D Virus and Mycoplasma bovis Coinfection in Calves: a Disease Severity Linked with Overexpression of IFN-γ as a Key Player of the Enhanced Innate Immune Response in Lungs. Kibenge FSB, editor. Microbiol Spectr. 9(3):e01690-21. https://doi.org/10.1128/spectrum.01690-21.
- Luan SJ, Sun YB, Wang Y, Sa RN, Zhang HF. 2019. Bacillus amyloliquefaciens spray improves the growth performance, immune status, and respiratory mucosal barrier in broiler chickens. Poult Sci. 98(3):1403–1409. https://doi.org/10.3382/ps/pey478.
- Macauley MS, Kawasaki N, Peng W, Wang S-H, He Y, Arlian BM, McBride R, Kannagi R, Khoo K-H, Paulson JC, et al. 2015. Unmasking of CD22 co-receptor on germinal center B-cells Z. J Biol Chem. 290(50):30066–30077. https://doi.org/10.1074/jbc.M115.691337.
- Mach N, Baranowski E, Nouvel LX, Citti C. 2021. The airway pathobiome in complex respiratory diseases: a perspective in domestic animals. Front Cell Infect Microbiol. 11:583600. https://doi.org/10.3389/fcimb.2021.583600.
- Mach N, Clark A. 2017. Micronutrient deficiencies and the human gut microbiota. Trends Microbiol. 25(8):607–610. https://doi.org/10.1016/j.tim.2017.06.004.
- Mach N, Plancade S, Pacholewska A, Lecardonnel J, Rivière J, Moroldo M, Vaiman A, Morgenthaler C, Beinat M, Nevot A, et al. 2016. Integrated mRNA and miRNA expression profiling in blood reveals candidate biomarkers associated with endurance exercise in the horse. Sci Rep. 6(1):22932. https://doi.org/10.1038/srep22932.
- Mahmmod YS, Correa-Fiz F, Aragon V. 2020. Variations in association of nasal microbiota with virulent and non-virulent strains of Glaesserella (Haemophilus) parasuis in weaning piglets. Vet Res. 51(1):7. https://doi.org/10.1186/s13567-020-0738-8.
- Malaker SA, Riley NM, Shon DJ, Pedram K, Krishnan V, Dorigo O, Bertozzi CR. 2022. Revealing the human mucinome. Nat Commun. 13(1):3542. https://doi.org/10.1038/s41467-022-31062-4.
- Malmuthuge N, Guan LL. 2021. Noncoding RNAs: regulatory molecules of host–microbiome crosstalk. Trends Microbiol. 29(8):713–724. https://doi.org/10.1016/j.tim.2020.12.003.
- Malmuthuge N, Liang G, Guan LL. 2019. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 20(1):172. https://doi.org/10.1186/s13059-019-1786-0.
- Man WH, De Steenhuijsen Piters WAA, Bogaert D. 2017. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 15(5):259–270. https://doi.org/10.1038/nrmicro.2017.14.
- Mariadassou M, Nouvel LX, Constant F, Morgavi DP, Rault L, Barbey S, Helloin E, Rué O, Schbath S, Launay F, et al. 2023. Microbiota members from body sites of dairy cows are largely shared within individual hosts throughout lactation but sharing is limited in the herd. Anim Microbiome. 5(1):32. https://doi.org/10.1186/s42523-023-00252-w.
- Martínez-Moliner V, Soler-Llorens P, Moleres J, Garmendia J, Aragon V. 2012. Distribution of genes involved in sialic acid utilization in strains of Haemophilus parasuis. Microbiology (Reading). 158(Pt 8):2117–2124. https://doi.org/10.1099/mic.0.056994-0.
- May M, Brown DR. 2009. Secreted sialidase activity of canine mycoplasmas. Vet Microbiol. 137(3–4):380–383. https://doi.org/10.1016/j.vetmic.2009.01.009.
- Mazel-Sanchez B, Yildiz S, Schmolke M. 2019. Ménage à trois: virus, host, and microbiota in experimental infection models. Trends Microbiol. 27(5):440–452. https://doi.org/10.1016/j.tim.2018.12.004.
- McMullen C, Alexander TW, Léguillette R, Workentine M, Timsit E. 2020. Topography of the respiratory tract bacterial microbiota in cattle. Microbiome. 8(1):91. https://doi.org/10.1186/s40168-020-00869-y.
- McMullen C, Orsel K, Alexander TW, van der Meer F, Plastow G, Timsit E. 2019. Comparison of the nasopharyngeal bacterial microbiota of beef calves raised without the use of antimicrobials between healthy calves and those diagnosed with bovine respiratory disease. Vet Microbiol. 231:56–62. https://doi.org/10.1016/j.vetmic.2019.02.030.
- Meldrum OW, Chotirmall SH. 2021. Mucus, microbiomes and pulmonary disease. Biomedicines. 9(6):675. https://doi.org/10.3390/biomedicines9060675.
- Meng F, Wu NH, Seitz M, Herrler G, Valentin-Weigand P. 2016. Efficient suilysin-mediated invasion and apoptosis in porcine respiratory epithelial cells after streptococcal infection under air-liquid interface conditions. Sci Rep. 6(1):26748. https://doi.org/10.1038/srep26748.
- Miao Y, Niu D, Wang Z, Wang J, Wu Z, Bao J, Hu W, Guo Y, Li R, Ishfaq M, et al. 2022. Mycoplasma gallisepticum induced inflammation-mediated Th1/Th2 immune imbalance via JAK/STAT signaling pathway in chicken trachea: involvement of respiratory microbiota. Vet Microbiol. 265:109330. https://doi.org/10.1016/j.vetmic.2021.109330.
- Much P, Winner F, Stipkovits L, Rosengarten R, Citti C. 2002. Mycoplasma gallisepticum: influence of cell invasiveness on the outcome of experimental infection in chickens. FEMS Immunol Med Microbiol. 34(3):181–186. (02)00378-4. https://doi.org/10.1016/S0928-8244.
- Muranaka M, Yamanaka T, Katayama Y, Hidari K, Kanazawa H, Suzuki T, Oku K, Oyamada T. 2011. Distribution of influenza virus sialoreceptors on upper and lower respiratory tract in horses and dogs. J Vet Med Sci. 73(1):125–127. https://doi.org/10.1292/jvms.10-0276.
- Nason R, Büll C, Konstantinidi A, Sun L, Ye Z, Halim A, Du W, Sørensen DM, Durbesson F, Furukawa S, et al. 2021. Display of the human mucinome with defined O-glycans by gene engineered cells. Nat Commun. 12(1):4070. https://doi.org/10.1038/s41467-021-24366-4.
- Ngunjiri JM, Taylor KJM, Abundo MC, Jang H, Elaish M, Kc M, Ghorbani A, Wijeratne S, Weber BP, Johnson TJ, et al. 2019. Farm stage, bird age, and body site dominantly affect the quantity, taxonomic composition, and dynamics of respiratory and gut microbiota of commercial layer chickens. Appl Environ Microbiol. 85(9):1–17. https://doi.org/10.1128/AEM.03137-18.
- Nicola I, Cerutti F, Grego E, Bertone I, Gianella P, D’Angelo A, Peletto S, Bellino C. 2017. Characterization of the upper and lower respiratory tract microbiota in Piedmontese calves. Microbiome. 5(1):152. https://doi.org/10.1186/s40168-017-0372-5.
- Oladunni FS, Horohov DW, Chambers TM. 2019. EHV-1: a constant threat to the horse industry. Front Microbiol. 10:2668. https://doi.org/10.3389/fmicb.2019.02668.
- Oliva J, Eichenbaum A, Belin J, Gaudino M, Guillotin J, Alzieu J-P, Nicollet P, Brugidou R, Gueneau E, Michel E, et al. 2019. Serological evidence of influenza D virus circulation among cattle and small ruminants in France. Viruses. 11(6):516. https://doi.org/10.3390/v11060516.
- Oliveira ED, Quaglio AEV, Magro DO, Di Stasi LC, Sassaki LY. 2023. Intestinal microbiota and miRNA in IBD: a narrative review about discoveries and perspectives for the future. Int J Mol Sci. 24(8):7176. https://doi.org/10.3390/ijms24087176.
- Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M, Boeren S, et al. 2017. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 12(3):e0173004. https://doi.org/10.1371/journal.pone.0173004.
- Pacholewska A, Mach N, Mata X, Vaiman A, Schibler L, Barrey E, Gerber V. 2016. Novel equine tissue miRNAs and breed-related miRNA expressed in serum. BMC Genomics. 17(1):831. https://doi.org/10.1186/s12864-016-3168-2.
- Pajic P, Shen S, Qu J, May AJ, Knox S, Ruhl S, Gokcumen O. 2022. A mechanism of gene evolution generating mucin function. Sci Adv. 8(34):eabm8757. https://doi.org/10.1126/sciadv.abm8757.
- Pant K, Yadav AK, Gupta P, Islam R, Saraya A, Venugopal SK. 2017. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol. 12:340–349. https://doi.org/10.1016/j.redox.2017.03.006.
- Paone P, Cani PD. 2020. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 69(12):2232–2243. https://doi.org/10.1136/gutjnl-2020-322260.
- Park GY, Yu HJ, Son JS, Park SJ, Cha H-J, Song KS. 2020. Specific bacteriophage of Bordetella bronchiseptica regulates B. bronchiseptica-induced microRNA expression profiles to decrease inflammation in swine nasal turbinate cells. Genes Genomics. 42(4):441–447. https://doi.org/10.1007/s13258-019-00906-7.
- Patel JG, Patel BJ, Patel SS, Raval SH, Parmar RS, Joshi DV, Chauhan HC, Chandel BS, Patel BK. 2018. Metagenomic of clinically diseased and healthy broiler affected with respiratory disease complex. Data Brief. 19:82–85. https://doi.org/10.1016/j.dib.2018.05.010.
- Pearce SC, Weber GJ, van Sambeek DM, Soares JW, Racicot K, Breault DT. 2020. Intestinal enteroids recapitulate the effects of short-chain fatty acids on the intestinal epithelium. PLoS One. 15(4):e0230231. https://doi.org/10.1371/journal.pone.0230231.
- Peng L, Li Z-R, Green RS, Holzman IR, Lin J. 2009. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in caco-2 cell monolayers. J Nutr. 139(9):1619–1625. https://doi.org/10.3945/jn.109.104638.
- Pereira FC, Wasmund K, Cobankovic I, Jehmlich N, Herbold CW, Lee KS, Sziranyi B, Vesely C, Decker T, Stocker R, et al. 2020. Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat Commun. 11(1):41467. https://doi.org/10.1038/s41467-020-18928-1.
- Pérez-Cobas AE, Rodríguez-Beltrán J, Baquero F, Coque TM. 2023. Ecology of the respiratory tract microbiome. Trends Microbiol. 31(9):972–984. https://doi.org/10.1016/j.tim.2023.04.006.
- Pirolo M, Espinosa-Gongora C, Bogaert D, Guardabassi L. 2021. The porcine respiratory microbiome: recent insights and future challenges. Anim Microbiome. 3(1):9. https://doi.org/10.1186/s42523-020-00070-4.
- Qin R, Mahal LK. 2021. The host glycomic response to pathogens. Curr Opin Struct Biol. 68:149–156. https://doi.org/10.1016/j.sbi.2020.12.011.
- Qin S, Ruan W, Yue H, Tang C, Zhou K, Zhang B. 2018. Viral communities associated with porcine respiratory disease complex in intensive commercial farms in Sichuan province, China. Sci Rep. 8(1):13341. https://doi.org/10.1038/s41598-018-31554-8.
- Rasaei D, Hosseinian SA, Asasi K, Shekarforoush SS, Khodakaram-Tafti A. 2023. The beneficial effects of spraying of probiotic Bacillus and Lactobacillus bacteria on broiler chickens experimentally infected with avian influenza virus H9N2. Poult Sci. 102(7):102669. https://doi.org/10.1016/j.psj.2023.102669.
- Reddy VRAP, Trus I, Nauwynck HJ. 2017. Presence of DNA extracellular traps but not MUC5AC and MUC5B mucin in mucoid plugs/casts of infectious laryngotracheitis virus (ILTV) infected tracheas of chickens. Virus Res. 227:135–142. https://doi.org/10.1016/j.virusres.2016.09.025.
- Reily C, Stewart TJ, Renfrow MB, Novak J. 2019. Glycosylation in health and disease. Nat Rev Nephrol. 15(6):346–366. https://doi.org/10.1038/s41581-019-0129-4.
- Ricci S, Petri RM, Pacífico C, Castillo-Lopez E, Rivera-Chacon R, Sener-Aydemir A, Reisinger N, Zebeli Q, Kreuzer-Redmer S. 2022. Characterization of presence and activity of microRNAs in the rumen of cattle hints at possible host-microbiota cross-talk mechanism. Sci Rep. 12(1):13812. https://doi.org/10.1038/s41598-022-17445-z.
- Ridley C, Thornton DJ. 2018. Mucins: the frontline defence of the lung. Biochem Soc Trans. 46(5):1099–1106. https://doi.org/10.1042/BST20170402.
- Rose MC, Voynow JA. 2006. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev. 86(1):245–278. https://doi.org/10.1152/physrev.00010.2005.
- Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, et al. 2014. Muc5b is required for airway defence. Nature. 505(7483):412–416. https://doi.org/10.1038/nature12807.
- Salem E, Dhanasekaran V, Cassard H, Hause B, Maman S, Meyer G, Ducatez MF. 2020. Global transmission, spatial segregation, and recombination determine the long-term evolution and epidemiology of bovine coronaviruses. Viruses. 12(5):534. https://doi.org/10.3390/v12050534.
- Samy A, Naguib MM. 2018. Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet Sci. 5(1):23. https://doi.org/10.3390/vetsci5010023.
- Schaefer A, Lai SK. 2022. The biophysical principles underpinning muco-trapping functions of antibodies. Hum Vaccin Immunother. 18(2):1939605. https://doi.org/10.1080/21645515.2021.1939605.
- Scher JU, Joshua V, Artacho A, Abdollahi-Roodsaz S, Öckinger J, Kullberg S, Sköld M, Eklund A, Grunewald J, Clemente JC, et al. 2016. The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome. 4(1):60. https://doi.org/10.1186/s40168-016-0206-x.
- Schnaar RL. 2015. Glycans and glycan-binding proteins in immune regulation: a concise introduction to glycobiology for the allergist. J Allergy Clin Immunol. 135(3):609–615. https://doi.org/10.1016/j.jaci.2014.10.057.
- Schulz BL, Sloane AJ, Robinson LJ, Prasad SS, Lindner RA, Robinson M, Bye PT, Nielson DW, Harry JL, Packer NH, et al. 2007. Glycosylation of sputum mucins is altered in cystic fibrosis patients. Glycobiology. 17(7):698–712. https://doi.org/10.1093/glycob/cwm036.
- Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology (Reading). 153(Pt 9):2817–2822. https://doi.org/10.1099/mic.0.2007/009480-0.
- Shamseer L, Moher D, Clarke M, Ghersi D, Liberati A, Petticrew M, Shekelle P, Stewart LA, the PRISMA-P Group. 2015. PRISMA-P (preferred reporting items for systematic review and meta-analysis protocols) 2015 checklist : recommended items to address in a systematic review protocol *. BMJ. 349(jan02 1):g7647–g7647. https://doi.org/10.1111/j.1532-950X.2010.00691.x.
- Sharp JM, Gilmour NJL, Rushton B. 1978. Ovine pneumonia. N Z Vet J. 26(6):165–165. https://doi.org/10.1080/00480169.1978.34527.
- Shipunov I, Kupaev V. 2022. Glycome assessment in patients with respiratory diseases. Trans Meta Syndrome Res. 5:10–15. https://doi.org/10.1016/j.tmsr.2022.02.001.
- Siddiqui S, Johansson K, Joo A, Bonser LR, Koh KD, Le Tonqueze O, Bolourchi S, Bautista RA, Zlock L, Roth TL, et al. 2021. Epithelial miR-141 regulates IL-13–induced airway mucus production. JCI Insight. 6(5):e139019. https://doi.org/10.1172/jci.insight.139019.
- Simon JC, Marchesi JR, Mougel C, Selosse MA. 2019. Host-microbiota interactions: from holobiont theory to analysis. Microbiome. 7(1):5. https://doi.org/10.1186/s40168-019-0619-4.
- Skovgaard K, Cirera S, Vasby D, Podolska A, Breum SØ, Dürrwald R, Schlegel M, Heegaard PMH. 2013. Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2). Innate Immun. 19(5):531–544. https://doi.org/10.1177/1753425912473668.
- Skrzypek K, Tertil M, Golda S, Ciesla M, Weglarczyk K, Collet G, Guichard A, Kozakowska M, Boczkowski J, Was H, et al. 2013. Interplay between heme oxygenase-1 and miR-378 affects non-small cell lung carcinoma growth, vascularization, and metastasis. Antioxid Redox Signal. 19(7):644–660. https://doi.org/10.1089/ars.2013.5184.
- Smirnova MG, Guo L, Birchall JP, Pearson JP. 2003. LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell Immunol. 221(1):42–49. https://doi.org/10.1016/S0008-8749.(03)00059-5.
- Sommer F, Nookaew I, Sommer N, Fogelstrand P, Bäckhed F. 2015. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 16(1):62. https://doi.org/10.1186/s13059-015-0614-4.
- Song D, Iverson E, Kaler L, Boboltz A, Scull MA, Duncan GA. 2022. MUC5B mobilizes and MUC5AC spatially aligns mucociliary transport on human airway epithelium. Sci Adv. 8(47):eabq5049. https://doi.org/10.1126/sciadv.abq5049.
- Stencel-Baerenwald JE, Reiss K, Reiter DM, Stehle T, Dermody TS. 2014. The sweet spot: defining virus–sialic acid interactions. Nat Rev Microbiol. 12(11):739–749. https://doi.org/10.1038/nrmicro3346.
- Su S, Fu X, Li G, Kerlin F, Veit M. 2017. Novel influenza D virus: epidemiology, pathology, evolution and biological characteristics. Virulence. 8(8):1580–1591. https://doi.org/10.1080/21505594.2017.1365216.
- Suzuki N, Abe T, Natsuka S. 2022. Structural analysis of N-glycans in chicken trachea and lung reveals potential receptors of chicken influenza viruses. Sci Rep. 12(1):2081. https://doi.org/10.1038/s41598-022-05961-x.
- Thai P, Loukoianov A, Wachi S, Wu R. 2008. Regulation of airway mucin gene expression. Annu Rev Physiol. 70(1):405–429. https://doi.org/10.1146/annurev.physiol.70.113006.100441.
- Thornton DJ, Rousseau K, McGuckin MA. 2008. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol. 70(1):459–486. https://doi.org/10.1146/annurev.physiol.70.113006.100702.
- Thu CT, Mahal LK. 2020. Sweet control: microRNA regulation of the glycome. Biochemistry. 59(34):3098–3110. https://doi.org/10.1021/acs.biochem.9b00784.
- Timoneda O, Núñez-Hernández F, Balcells I, Muñoz M, Castelló A, Vera G, Pérez LJ, Egea R, Mir G, Córdoba S, et al. 2014. The role of viral and host MicroRNAs in the Aujeszky’s disease virus during the infection process. PLoS One. 9(1):e86965. https://doi.org/10.1371/journal.pone.0086965.
- Timsit E, Workentine M, Crepieux T, Miller C, Regev-Shoshani G, Schaefer A, Alexander T. 2017. Effects of nasal instillation of a nitric oxide-releasing solution or parenteral administration of tilmicosin on the nasopharyngeal microbiota of beef feedlot cattle at high-risk of developing respiratory tract disease. Res Vet Sci. 115:117–124. https://doi.org/10.1016/j.rvsc.2017.02.001.
- Umar S, Guerin JL, Ducatez MF. 2016. Low pathogenic avian influenza and coinfecting pathogens: a review of experimental infections in avian models. Avian Dis. 61(1):3–15. https://doi.org/10.1637/11514-101316-review.
- van Leenen K, Van Driessche L, De Cremer L, Masmeijer C, Boyen F, Deprez P, Pardon B. 2020. Comparison of bronchoalveolar lavage fluid bacteriology and cytology in calves classified based on combined clinical scoring and lung ultrasonography. Prev Vet Med. 176:104901. https://doi.org/10.1016/j.prevetmed.2020.104901.
- Varki A. 2007. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 446(7139):1023–1029. https://doi.org/10.1038/nature05816.
- Varki A. 2008. Sialic acids in human health and disease. Trends Mol Med. 14(8):351–360. https://doi.org/10.1016/j.molmed.2008.06.002.
- Vayssier-Taussat M, Kazimirova M, Hubalek Z, Hornok S, Farkas R, Cosson J-F, Bonnet S, Vourch G, Gasqui P, Mihalca AD, et al. 2015. Emerging horizons for tick-borne pathogens: from the “one pathogen-one disease” vision to the pathobiome paradigm. Future Microbiol. 10(12):2033–2043. https://doi.org/10.2217/fmb.15.114.
- Vlasak R, Luytjes W, Spaan W, Palese P. 1988. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. 85(12):4526–4529. https://doi.org/10.1073/pnas.85.12.4526.
- Wallace LE, Liu M, van Kuppeveld FJM, de Vries E, de Haan CAM. 2021. Respiratory mucus as a virus-host range determinant. Trends Microbiol. 29(11):983–992. https://doi.org/10.1016/j.tim.2021.03.014.
- Walther T, Karamanska R, Chan RWY, Chan MCW, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, et al. 2013. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 9(3):e1003223. https://doi.org/10.1371/journal.ppat.1003223.
- Wang J, Ishfaq M, Li J. 2021. Lactobacillus salivarius ameliorated Mycoplasma gallisepticum-induced inflammatory injury and secondary Escherichia coli infection in chickens: involvement of intestinal microbiota. Vet Immunol Immunopathol. 233:110192. https://doi.org/10.1016/j.vetimm.2021.110192.
- Wang Q, Cai R, Huang A, Wang X, Qu W, Shi L, Li C, Yan H. 2018. Comparison of oropharyngeal microbiota in healthy piglets and piglets with respiratory disease. Front Microbiol. 9:3218. https://doi.org/10.3389/fmicb.2018.03218.
- Wang T, He Q, Yao W, Shao Y, Li J, Huang F. 2019. The variation of nasal microbiota caused by low levels of gaseous ammonia exposure in growing pigs. Front Microbiol. 10:1083. https://doi.org/10.3389/fmicb.2019.01083.
- Wang X, Chen H, Liu J, Gai L, Yan X, Guo Z, Liu F. 2021. Emerging advances of non-coding RNAs and competitive endogenous RNA regulatory networks in asthma. Bioengineered. 12(1):7820–7836. https://doi.org/10.1080/21655979.2021.1981796.
- Wasik BR, Barnard KN, Parrish CR. 2016. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 24(12):991–1001. https://doi.org/10.1016/j.tim.2016.07.005.
- Wheeler KM, Cárcamo-Oyarce G, Turner BS, Dellos-Nolan S, Co JY, Lehoux S, Cummings RD, Wozniak DJ, Ribbeck K. 2019. Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat Microbiol. 4(12):2146–2154. https://doi.org/10.1038/s41564-019-0581-8.
- Wypych TP, Wickramasinghe LC, Marsland BJ. 2019. The influence of the microbiome on respiratory health. Nat Immunol. 20(10):1279–1290. https://doi.org/10.1038/s41590-019-0451-9.
- Xia B, Royall JA, Damera G, Sachdev GP, Cummings RD. 2005. Altered O-glycosylation and sulfation of airway mucins associated with cystic fibrosis. Glycobiology. 15(8):747–775. https://doi.org/10.1093/glycob/cwi061.
- Yitbarek A, Astill J, Hodgins DC, Parkinson J, Nagy É, Sharif S. 2019. Commensal gut microbiota can modulate adaptive immune responses in chickens vaccinated with whole inactivated avian influenza virus subtype H9N2. Vaccine. 37(44):6640–6647. https://doi.org/10.1016/j.vaccine.2019.09.046.
- Yitbarek A, Taha-Abdelaziz K, Hodgins DC, Read L, Nagy É, Weese JS, Caswell JL, Parkinson J, Sharif S. 2018. Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci Rep. 8(1):13189. https://doi.org/10.1038/s41598-018-31613-0.
- Yu X, Dang VT, Fleming FE, von Itzstein M, Coulson BS, Blanchard H. 2012. Structural basis of rotavirus strain preference toward N-acetyl- or N-glycolylneuraminic acid-containing receptors. J Virol. 86(24):13456–13466. https://doi.org/10.1128/JVI.06975-11.
- Zeineldin M, Elolimy AA, Barakat R. 2020. Meta-analysis of bovine respiratory microbiota: link between respiratory microbiota and bovine respiratory health. FEMS Microbiol Ecol. 96(8):fiaa127. https://doi.org/10.1093/femsec/fiaa127.
- Zeineldin M, Lowe J, Aldridge B. 2019. Contribution of the mucosal microbiota to bovine respiratory health. Trends Microbiol. 27(9):753–770. https://doi.org/10.1016/j.tim.2019.04.005.
- Zeineldin M, Lowe J, de Godoy M, Maradiaga N, Ramirez C, Ghanem M, Abd El-Raof Y, Aldridge B. 2017. Disparity in the nasopharyngeal microbiota between healthy cattle on feed, at entry processing and with respiratory disease. Vet Microbiol. 208:30–37. https://doi.org/10.1016/j.vetmic.2017.07.006.
- Zhang M, Hill JE, Fernando C, Alexander TW, Timsit E, van der Meer F, Huang Y. 2019. Respiratory viruses identified in western Canadian beef cattle by metagenomic sequencing and their association with bovine respiratory disease. Transbound Emerg Dis. 66(3):1379–1386. https://doi.org/10.1111/tbed.13172.
- Zhang Y, Wang L, Qiu L, Pan R, Bai H, Jiang Y, Wang Z, Bi Y, Chen G, Chang G, et al. 2019. Expression patterns of novel circular RNAs in chicken cells after avian leukosis virus subgroup J infection. Gene. 701:72–81. https://doi.org/10.1016/j.gene.2019.03.030.
- Zhong T, Perelman JM, Kolosov VP, Zhou X. 2011. MiR-146a negatively regulates neutrophil elastase-induced MUC5AC secretion from 16HBE human bronchial epithelial cells. Mol Cell Biochem. 358(1–2):249–255. https://doi.org/10.1007/s11010-011-0975-2.