Abstract
Crisp grass carp is a type of grass carp (Ctenopharyngodon idella) that has been fed faba beans. Flesh quality and expression of muscle quality-related genes in muscle of grass carp were evaluated. Compared with ordinary grass carp, 1901 differentially expressed genes (DEGs) were up-regulated and 1628 DEGs were down-regulated in crisp grass carp. The GO analysis results showed that the DEGs were enriched in ribosomes and proteins. Kyoto Encyclopaedia of genes and genomes (KEGG) pathway analysis results showed that amino acid metabolism was closely related to meat quality. COX3, ACADS, ACAT1, ECHS1, EHHADH, HADHAA, HADH, HMGCL, MCCC1 and MCCC2 were the top 10 hub genes most closely associated with other nodes and showed a strong positive correlation. Hardness was positively correlated with the COX3, ECHS1, ACADS, ACAT1, HADHA, HADH, HMGCL and MCCC1. These results have been demonstrated to play important roles in clarifying the molecular mechanism of muscle firmness increase in crisp grass carp.
Highlights
Feeding faba bean meal improved texture parameters of grass carp.
A more comprehensive crisp grass carp transcriptome database was established.
Several amino acid metabolism pathway that were closely related to meat quality were identified.
Introduction
Grass carp (Ctenopharyngodon idellus) is a kind of freshwater fish which is widely farmed in China and has great economic importance (Yang et al. Citation2020). Grass carp is well known for excellent adaptability, rapid growth and large size, and it is one of the ‘Four Domesticated Fish’ in China (Liu et al. Citation2013). Through digestion and absorption, it can convert plant protein into animal protein directly (Lu et al. Citation2020). Grass carp is one of the most productive freshwater fish in China, which is also the species fish with the highest yield in freshwater fish farming worldwide (Rao and Su Citation2015; Wu et al. Citation2022).
Fish is an essential portion of the human diet because it is an excellent source of protein and unsaturated fatty acids, both of which are thought to be beneficial to human health. While breeding output is rising, it frequently appears that meat quality is declining, which has a significant negative impact on the meat’s economic value. Numerous studies have been attempted to enhance the quality of the meat from farmed grass carp (Yu et al. Citation2020; Yang et al. Citation2022; Li et al. Citation2023). Among them, feeding grass carp with faba bean to embrittle its meat quality is an innovative method that enhances the meat quality of grass carp (Zhou et al. Citation2019). Grass carp raised by feeding faba bean are called crisp grass carp (Lin et al. Citation2009). Studies have shown that the muscle characteristics of crisp grass carp have greatly changed compared with ordinary grass carp, in which the hardness, chewiness and springiness have increased significantly, the diameter of muscle fibres has decreased, and the density of muscle fibres has increased (Lin et al. Citation2009), which has increased its added value. Faba beans are also known to increase muscle hardness in fish such as Dicentrarchus labrax (Adamidou et al. Citation2009) and Oreochromis niloticus (Li et al. Citation2023). Fava beans also affect the fatty acid content of fish, increasing fat deposits in the viscera (Tian et al. Citation2019).
In the previous studies of crisp grass carp muscle, it has been found that the chewiness, flesh hardness, adhesiveness and elasticity of grass carp were significantly increased by feeding the fish faba beans (Yang et al. Citation2015), and the amino acid and fatty acid composition were influenced by fava bean in grass carp (Jiang et al. Citation2020; Li et al. Citation2020; Xu et al. Citation2020). The amount of collagen was another factor affecting the firmness of fish meat. A higher content of collagen produces a firmer raw fish meat, and the increase in the expression of type I collagen in crisp grass carp is higher than those of grass carp (Yu et al. Citation2014, Citation2019). However, long-term intake of whole fava bean can lead to poor palatability of crisp grass carp, oxidative stress, intestinal inflammation and decreased growth rate (Yu et al. Citation2014, Citation2017; Ma et al. Citation2020), and the maximal proportion of dietary protein replacement with faba bean meal was 70% based on the flesh quality of grass carp (Fu et al. Citation2022). Therefore, it is expedient to analyse systematically muscle firmness increase of crisp grass carp in the gene levels. Understanding the impact of faba beans on skeletal muscle quality is important for optimising feed used for high-quality crisp grass carp production.
Improving the muscle mass of fish is a major goal of aquaculture. Transcriptome analysis can accurately, efficiently and quickly obtain transcriptome sequence information and expression of specific cells or tissues in a certain state, as well as analyse changes in gene expression (Azaman et al. Citation2019; Gill and Dhillon Citation2022), which provides an effective way to explore the molecular mechanism of faba bean regulating grass carp meat quality. This study intends to perform transcriptome sequencing on the muscle tissue of crisp grass carp and ordinary grass carp and to screen out differentially expressed genes (DEGs) and signalling pathways related to muscle, so as to lay a foundation for analysing the molecular mechanism of faba bean regulating the development of grass carp meat quality.
Materials and methods
Tissue collection of experimental fish
Healthy grass carp were purchased from an aquaculture farm in Zunyi, Guizhou Province, China. The fish were first temporarily cultured in a cement pond (5 m × 5 m × 1.5 m) for 1 week and the feed amount for each day was 2–3% of fish weight. A total of 180 fish with initial weight of 768 ± 75 g were randomly divided into crisp grass carp (group A) and ordinary grass carp groups (group B), with three replicates each group. They were cultured in six cement ponds (2 × 2 × 1.5 m), with 30 fish in each pond. Crisp grass carp were fed solely with whole faba beans, the feed was soaked in about 0.15% salt water for 24 h, and then soaked in water for 12 h until the broad beans were opened after the germ. The single feeding amount of broad bean accounted for 2–3% of the body weight of fish. The ordinary grass carp were fed with commercial diet (crude protein: 329.9 g kg−1; crude lipid: 43.8 g kg−1; Tongwei, Chengdu, China). The fish were fed twice per day (at 8:00 and 17:00). The water temperature was kept at 25–30 °C, pH was 6.5–7.5, and dissolved oxygen was above 5.0 mg/L. The final weights of crisp grass carp and ordinary grass carp were 1992 ± 125 g and 2457 ± 132 g after 120 days, respectively. One fish was randomly selected from each pond to collect its muscle tissue, which was placed at −80 °C until RNA extraction. The procedure in this experiment was approved by the Ethics Committee of Experimental Animal of Zunyi Normal College (protocol number: ZUNSHIFA[2018]08).
Determination of muscle physical parameters and texture characteristics
Five fish from each group were used for determination of muscle physical and texture parameters. The muscle pH was analysed following the method reported by Knowles et al. (Citation2007). Cooking loss was measured as follows: 2–3 g of dorsal muscle samples were dried with filter papers and weighed (weight was W0), and then heated for 1 min in a pan of boiling water. The samples were weighed again (weight was W1) after cooling to room temperature and removing surface water. The calculation formula was as follows: cooking loss (%) = 100 × (W0 – W1)/W0. The texture of the dorsal muscle, including hardness, springiness, chewiness and cohesiveness, was performed with the texture profiles analysis (TPA) using a Universal TA device (Tengba, Shanghai, China). Test conditions were as follows: a 25 mm × 25 mm flat-bottomed cylindrical probe, a compression ratio of 30%, a test speed of 1 mm/s and a post-test speed of 1 mm/s with the staying time of 2 s. Test conditions of shear force were as follows: compression distance of 20 mm, a test speed of 1 mm/s and a post-test speed of 1 mm/s with a staying time of 2 s.
RNA extraction and sequencing
Total RNA was isolated using the Trizol Reagent (Invitrogen Life Technologies, Carlsbad, CA), after which the concentration, quality and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). Three micrograms of RNA were used as input material for the RNA sample preparations. The mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. The cDNA was synthesised using the mRNA fragment as templates. To select cDNA fragments of the preferred 400–500 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, CA). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using Illumina PCR Primer Cocktail in a 15 cycle PCR reaction. Products were purified (AMPure XP system, Beckman Coulter, Beverly, CA) and quantified using the Agilent high sensitivity DNA assay on a Bioanalyser 2100 system (Agilent, Santa Clara, CA). The sequencing library was then sequenced on NovaSeq 6000 platform (Illumina, San Diego, CA).
Transcriptome assembly and reads mapping
Samples are sequenced on the platform to get image files, which are transformed by the software of the sequencing platform, and the original data in FASTQ format (Raw Data) are generated. Sequencing data contain a number of connectors, low-quality Reads, so we use Cutadapt (v1.15) software to filter the sequencing data to get high quality sequence (Clean Data) for further analysis. The reference genome and gene annotation files were downloaded from genome website. The filtered reads were mapping to the reference genome using HISAT2 v2.0.5.
Differential expression analysis
HTSeq (0.9.1) statistics was used to compare the Read Count values on each gene as the original expression of the gene, and then used FPKM to standardise the expression. Then difference expression of genes was analysed by DESeq (1.30.0) with screened conditions as follows: expression |log2FoldChange| >1, significant p value <.05. At the same time, we used R language Pheatmap (1.0.8) software package to perform bi-directional clustering analysis of all different genes of samples. We got the heatmap according to the expression level of the same gene in different samples and the expression patterns of different genes in the same sample with Euclidean method to calculate the distance and Complete Linkage method to cluster.
GO and KEGG enrichment analysis
All the genes to terms in the Gene Ontology database were mapped and the numbers of differentially enriched genes in each term were calculated. Using topGO to perform GO enrichment analysis on the differential genes, calculate p value by hypergeometric distribution method (the standard of significant enrichment is p value <.05), and find the GO term with significantly enriched differential genes to determine the main biological functions performed by differential genes. ClusterProfiler (3.4.4) software was used to carry out the enrichment analysis of the Kyoto Encyclopaedia of genes and genomes (KEGG) pathway of differential genes, focusing on the significant enrichment pathway with p value <.05.
Interaction analysis of differential gene protein network
The STRING database (https://string-db.org/) is used for protein interaction analysis to reveal the relationship between target genes. The protein–protein interaction (PPI) network of the DEGs was constructed based on the Search Tool for the Retrieval of Interacting Genes (STRING) database22 (STRING, version 9.1, http://string91.embl.de/), and predicted by the use of Cytoscape23 (version 3.0; http://cytoscape.org/). The hub genes were then selected with a connectivity degree 10 after calculating the degree of each node. Module analysis of the PPI network was performed with the parameters of minimum size >4 and minimum density <0.05 using ClusterONE.
Correlation analysis and qRT-PCR validation
To validate the results of the transcriptome data, top 10 hub genes were randomly selected and their mRNA levels were measured by qRT-PCR. Specific primer sequences for qRT-PCR were as follows: peroxisomal acyl-coenzyme A oxidase 3 (COX3, 5F: CCAGTCCCTCACCCTAACAA; R: GTGGTGTTCGGAGGTGAAGT), enoyl-CoA hydratase (ECHS1, 5F: TAAGGCCTTGGACACCTTTG; R: TTTGTTCCAGTGTGCCAAAA), short-chain specific acyl-CoA dehydrogenase (ACADS, 5F: TCGACAGGAGTCATCGTCAG; R: TTGAGAACCCATTCGTTTCC), acetyl-CoA acetyltransferase (ACAT1, 5F: TGTCATCGTCAGTGCTGTCA; R: AGCTTGTCTGGTTGGTGCTT), peroxisomal bifunctional enzyme (EHHADH, 5F: GGGTGTGTAGACCAGCCACT; R: ATCCTCTTCCCAAGGCTCAT), trifunctional enzyme subunit alpha (HADHA, 5F: TGCCATCAAGAGTGCTGTTC; R: GGCTACAATGGGTTTGGATG), trifunctional enzyme subunit alpha (HADH, 5F: TAAATCTGCGAAGGGGATTG; R: ACAATAGCCTCCACCACCAG), hydroxymethylglutaryl-CoA lyase (HMGCL, 5F: CTCGGTGGAGGAGAGTCTTG; R: TGTCACCCAATGAAACCTCA), methylcrotonyl-CoA carboxylase subunit alpha (MCCC1, 5F: CAACATCGACTTCCTGCTGA; R: TCGCTGGTCTGCTCTCTGTA) and methylcrotonyl-CoA carboxylase beta chain (MCCC2, 5F: AGCCAGAGTTCGTCCTTTGA; R: GTGCTTGCATTCGCTCATAA). The total mRNA of the samples was extracted using the Trizol Kit. PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Kusatsu, Japan) was then used for cDNA synthesis based on manuals. qRT-PCR was conducted with a Roche Step One real-time PCR system (Roche, Indianapolis, IN). PCR amplification of all samples was performed in triplicate. β-Actin (5F: TGCCATGTATGTGGCCATCC; R: TCTTTCGGCTGTGGTGGTGA) was used as the normalisation control to calculate the relative expression levels (Li et al. 2019). The melting curve was analysed to confirm the target specificity. The 2−ΔΔCT method was used to calculate the relative gene expression values.
Results
Muscle physical parameters and texture characteristics
In this study, the muscle physical parameters and texture characteristics for ordinary grass carp and crisp grass carp are given in Table . Lower water holding capacity was observed in crisp grass carp group when compared with ordinary grass carp group (p < .05). Hardness, springiness and gumminess in crisp grass carp group were significantly (p < .05) higher compared with ordinary grass carp. Besides, muscle pH, cooking loss, chewiness and cohesiveness showed no obvious difference between the two groups (p > .05).
Table 1. Muscle physical and texture parameters of grass carp after 120-days feeding experiment.
Transcriptome sequencing and assembly results
In this study, a total of 40,564,018–44,425,106 raw reads and 37,910,970–41,601,388 clean reads were obtained through transcriptome analysis of six muscle tissue samples. The alignment rate of clean reads from ordinary grass crap and crisp grass crap mapped to the reference genome ranged from 94.28% to 95.14%. The Q20 is higher than 98.06% and the Q30 is higher than 94.49% (Table ). The generated sequence data have been submitted to the NCBI SRA database (accession number: PRJNA1053680).
Table 2. Summary statistics of transcriptome sequencing from muscle tissue between ordinary grass carp and the crisp carp group.
Differentially expressed gene identification and analysis
Gene expression was calculated according to the FPKM algorithm. Differential analysis of gene expression was performed using DESeq, and screening was performed according to |log2FoldChange| >1 and p value < .05. There were 3529 DEGs in crisp grass carp (group A) compared with ordinary grass carp groups (group B). Among them, 1901 differential genes were down-regulated, and 1628 differential genes were up-regulated (Figure ). In the DEGs of the top 20 up-regulated, large neutral amino acids transporter small subunit 4 (SLC43A2) is the gene with the most significant difference. In the DEGs of the top 20 down-regulated, l-lactate dehydrogenase A (LDHA) is the gene with the most significant difference. Among all DEGs, some genes related to muscle growth and development were also screened out, such as insulin-like growth factor-binding protein 2, myosin heavy chain (Table ). Differentially expressed genes (DEGs) identified in the muscle tissue between ordinary grass carp and the crisp carp group (Top20) are presented in Figure . GO enrichment analysis showed that DEGs were mainly enriched in collagen type I trimer, l-amino acid transmembrane transporter activity, carboxylic acid transmembrane transporter activity, amino acid transmembrane transporter activity, l-leucine transmembrane transporter activity, neutral amino acid transmembrane transporter activity, neutral amino acid transport, amino acid transport, l-alpha-amino acid transmembrane transport, branched-chain amino acid transmembrane transporter activity, leucine import across plasma membrane, amino acid transmembrane transport and collagen metabolic process. KEGG enrichment analysis showed that DEGs were mainly enriched in protein digestion and absorption, cysteine and methionine metabolism, glycolysis/gluconeogenesis, biosynthesis of amino acids, adipocytokine signalling pathway, tryptophan metabolism and pyruvate metabolism.
Figure 1. Volcano map and heat map of clustering of differentially expressed genes in Ctenopharyngodon idellus.
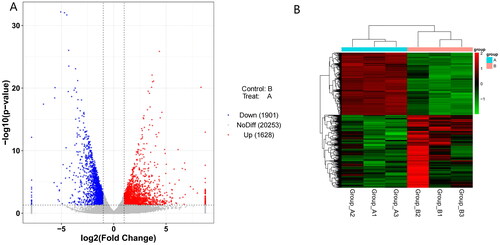
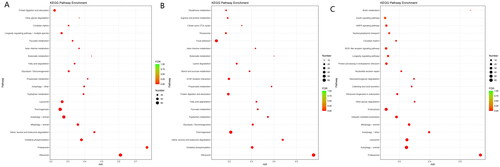
Figure 2. Classify and analysis according to gene function. (A) GO enrichment of top 20 up-regulated and down-regulated DEGs. (B) KEGG enrichment of top 20 up-regulated and down-regulated DEGs.
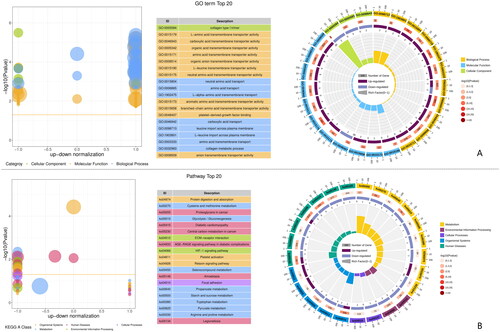
Table 3. Differentially expressed genes (DEGs) identified in the muscle tissue between ordinary grass carp and the crisp carp group (Top20).
GO enrichment analysis of DEGs
The results of GO enrichment analysis showed that DEGs were mainly enriched in 5484 GO terms of biological process (BP), molecular function (MF) and cellular component (CC), among which BP was the most prevalent (3493). In the BP, the DEGs are mainly involved in BP and cellular process, containing 1072 and 916 DEGs, respectively. In the MF, the DEGs involved in the MF process are the most, including 1030 DEGs. In the CC, the DEGs are mainly involved in the process of CC and cellular anatomical entity, including 967 and 927 DEGs, respectively.
GO enrichment analysis showed that the top 20 terms enriched DEGs were enriched in proteins and ribosomes (Figure ). GO enrichment showed that up-regulated genes were significantly enriched in the ribosomes, including ribonucleoprotein complex (GO:1990904), large ribosomal subunit (GO:0015934), small ribosomal subunit (GO:0015935), cytosolic small ribosomal subunit (GO:0022627), cytosolic large ribosomal subunit (GO:0022625), ribosomal subunit (GO:0044391), cytosolic ribosome (GO:0022626), ribosome (GO:0005840) and structural constituent of ribosome (GO:0003735). Protein-related GO terms include peptide biosynthetic process (GO:0043043), peptide metabolic process (GO:0006518) and translation (GO:0006412) (Figure ).
Figure 3. (A) GO enrichment of DEGs. (B) GO enrichment of up-regulated DEGs. (C) GO enrichment of down-regulated DEGs.
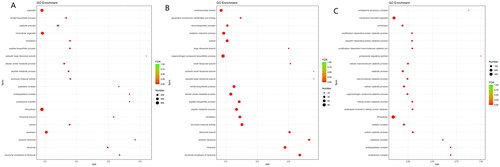
GO enrichment analysis showed that down-regulated genes were significantly enriched in the protein catabolism, including proteasome accessory complex (GO:0022624), proteolysis (GO:0006508), modification-dependent protein catabolic process (GO:0019941), ubiquitin-dependent protein catabolic process (GO:0006511), proteasome regulatory particle (GO:0005838), organonitrogen compound catabolic (GO:1901565), process cellular protein catabolic process (GO:0044257), proteolysis involved in cellular protein catabolic (GO:0051603), protein catabolic process (GO:0030163), peptidase complex (GO:1905368), endopeptidase complex (GO:1905369) and proteasome complex (GO:0000502) (Figure ).
KEGG enrichment analysis of DEGs
KEGG enrichment analysis showed that DEGs in muscle tissue were enriched into 252 known KEGG pathways, of which 45 signalling pathways were significantly enriched (p < .05).
The top 20 KEGG pathways enriched by DEGs are shown in Figure , and nine of the 20 top pathways were related to metabolism of energy, protein, amino acid and fatty acid. The top 20 KEGG pathways enriched by up-regulated differential genes are shown in Figure . There were six KEGG pathways related to amino acid metabolism, including arginine and proline metabolism, beta-alanine metabolism, lysine degradation, glutathione metabolism, tryptophan metabolism and valine, leucine and isoleucine degradation. Ribosome is also an upregulated GO term. The top 20 KEGG pathways enriched by down-regulated differential genes are shown in Figure . There were two pathways related to protein, including protein processing in endoplasmic reticulum and ubiquitin mediated proteolysis.
PPI network construction
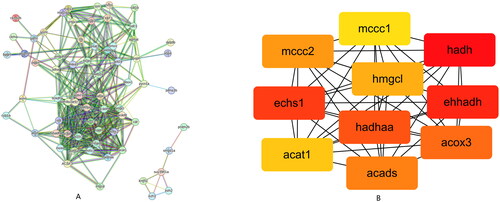
The PPI network with 72 nodes was constructed by analysing DEGs associated to differential amino acid metabolic pathways using the STRING database (Figure ). COX3, ECHS1, ACADS, ACAT1, EHHADH, HADHA, HADH, HMGCL, MCCC1 and MCCC2 were the top 10 hub genes most closely associated with other nodes, according to further research with cytoHubba (Figure ).
Correlation analysis and qRT-PCR validation
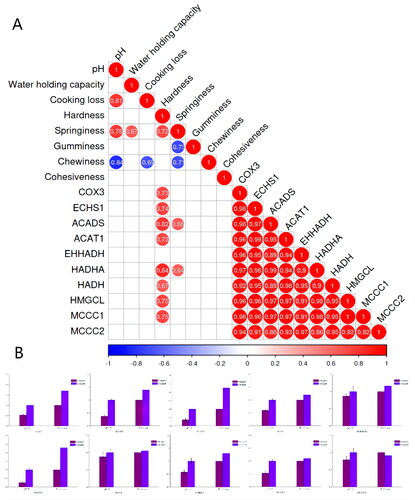
The correlations of top 10 hub genes with meat quality are shown in Figure . Hardness was positively correlated with the COX3, ECHS1, ACADS, ACAT1, HADHA, HADH, HMGCL and MCCC1. There is a strong positive correlation among the top 10 hub genes. To validate the RNA-Seq results obtained, 10 genes were randomly selected for validation by qRT-PCR. These genes that were involved in the amino acid metabolism were closely related to meat quality. As shown in Figure , the qPCR expression patterns of the selected genes agreed with the results of RNA-Seq analysis, indicating the reliability and accuracy of the RNA-Seq method used in the present study.
Discussion
Changes in muscle texture are generally regarded as an important indicator of the ‘embrittlement’ of crisp grass carp, especially the chewiness of muscles, which are an important factor for the ‘crispy’ of muscles. Feeding faba beans is an effective way to improve the meat quality of grass carp. Our results also showed that some texture parameters of crisp grass carp, such as the hardness, springiness and gumminess were significantly improved, while the water holding capacity in muscle was reduced. These results indicated that feeding faba bean was an effective way for improving muscle texture of grass carp.
SLC43A2 mediates the transport of leucine, phenylalanine, valine and methionine, which is critical for early development in animal (Guetg et al. Citation2015). LDHA can catalyse the conversion of l-lactic acid into pyruvate, which is closely related to the generation of ATP (Feng et al. Citation2018). Myosin, which contains components like MHC and MLC, is one of the primary structural and functional proteins that make up skeletal muscle and cardiac muscle (Agarwal et al. Citation2020). The size, quantity and type of muscle fibres are all influenced by MHC during the development of skeletal muscle. In the present study, the expression of 1628 genes in crispy grass carp was significantly up-regulated, while the expression of 1901 genes was significantly down-regulated. In the DEGs of the top 20 up-regulated, SLC43A2 is the most significant difference. In the DEGs of the top 20 down-regulated, LDHA is the most significant difference. Genes of type I collagen and type II collagen were once considered tightly related to muscle-hardening in grass carp fed with faba bean (Yu et al. Citation2019), and we have identified these genes in the current study. These DEGs may be related to the muscle growth and development of grass carp, which is worthy of further digging and research.
In this study, eight of the top 20 up-regulated GO terms were related to ribosomes. In fact, ribosomes are the main place for cells to synthesise proteins, and the process of RNA translation into proteins is completed in ribosomes (Zimmermann et al. Citation2016), whereas eukaryotic 80S ribosomes are composed of 40S small and 60S large subunits. The small subunit decodes transcripts by promoting base-pairing between codons within an mRNA and their corresponding transfer RNAs (tRNAs) (Buszczak Citation2023). The large subunit contains the peptidyl transferase centre which catalyses peptide bond formation between amino acids (Buszczak Citation2023). Most of the protein in skeletal muscle consists of the contractile proteins actin and myosin. The myocyte hypertrophy is independent of the proliferation of satellite cells and myonuclear accretion, in contrast to long-term remodelling and maintenance of skeletal muscle homeostasis, which appear to require satellite cells together with their influence on non-skeletal muscle cells (Fry et al. Citation2017). Our results showed that ribosome was the most DEGs among the top 20 up-regulated KEGG pathways, suggesting that the muscle cell coordinates its anabolic and nonanabolic processes in a complex and sophisticated manner, and ribosome biogenesis is important for hypertrophic growth of skeletal muscle. At the same time, translation and transcription are clearly linked and preferably synchronised during this process, probably to optimise the response to energy demands and the potential for growth and muscle adaptation (von Walden Citation2019). In addition, ribosomal proteins have been shown to influence cell differentiation, proliferation and death as well as play an essential component in muscle growth (Genuth and Barna Citation2018), and our results suggest that ribosomal proteins play an important role in maintaining the body’s normal life activities, and the content of amino acids is related to muscle nutrition and quality.
Meat quality, including amino acid content, is an important factor affecting meat flavour and consumer acceptance (Zhang et al. Citation2022). Amino acid concentration is an essential indicator for assessing the quality of meat and is crucial in enhancing meat flavour (Huo et al. Citation2021; San et al. Citation2021). It has been reported that in farmed fish, the composition of tissue amino acid is greatly affected by dietary nutrients. This is consistent with those previous studies, among the up-regulated pathways caused by feeding faba beans, there are six KEGG pathways related to amino acid metabolism, indicating that arginine, proline, β-alanine, lysine, tryptophan, leucine and isoleucine acid play an important role in improving grass carp meat quality. Some KEGG analysis results of this study are similar to those of other fishes, such as glycolysis/gluconeogenesis (Li et al. Citation2017), arginine and proline metabolism (Canosa and Bertucci Citation2020). These KEGG pathways, such as glycolysis, which produces ATP from glycogen, and gluconeogenesis, which produces glucose from non-sugar organic matter, are all connected to muscle growth and development. In a refeeding experiment with rainbow trout, certain genes involved in glycolysis and gluconeogenesis were discovered (Johansen and Overturf Citation2006). Fatty acids are one of the primary energy sources for organisms since they may be oxidised and broken down to produce a substantial amount of energy (De Carvalho and Caramujo Citation2018). Proline and arginine are two amino acids that can be consumed to increase muscle growth and decrease extra fat (Wu Citation2013). In a certain range, increasing the level of faba beans in the feed increased the content of total saturated fatty acids, total monounsaturated fatty acids, polyunsaturated fatty acids and n-3 polyunsaturated fatty acids in the grass carp muscles. Exceeding the range would reduce its deposition in the muscles. The relationship between these KEGG pathways and muscle growth and development in fish can be further investigated in the future.
Studies have shown that material and energy metabolism can affect biological growth (Canosa and Bertucci Citation2020). This study also confirmed this point: the GO and KEGG enrichment results of DEGs mostly focused on material and energy metabolism. Six KEGG pathways involved in amino acid metabolism were shown to be up-regulated by feeding faba beans in the KEGG enrichment results, and there were alterations in the glycolysis/gluconeogenesis pathway. Fast-growing grass carp have been found to have dramatically increased expression of DEGs enriched in metabolic activities (Lu et al. Citation2020). These results indicated that the addition of faba beans had an important impact on the amino acid metabolism and energy metabolism of grass carp.
In this study, the construction of PPI network and the identification of hub genes were carried out for the DEGs enriched in different KEGG pathways linked to amino acid metabolism, fatty acid metabolism and immunity. A total of 10 hub genes were discovered, including ACOX3, ACADS, ACAT1, ECHS1, EHHADH, HADHA, HADH, HMGCL, MCCCC1 and MCCC2. During the feeding period of grass carp fed with faba beans, the immunity of crisp grass carp was reduced and its mortality increased. In this study, we found that many down-regulated genes in crisp grass carp were clustered into amino acid catabolism and immune-related category. For instance, down regulation of ECHS1 induces fatty acid and branched-chain amino acid BCAA accumulation (Qu et al. Citation2020). MCCC1 (Zhao et al. Citation2020) is a key enzyme involved in the BP of amino acid metabolism, which is related to the catabolism of valine and leucine. MCCC2 (Chen et al. Citation2021) also functions to regulate leucine metabolism.
ACOX3 has a role in regulating lipid homeostasis (Madureira et al. Citation2016). ACADS also regulate fatty acid metabolism and regulate energy homeostasis (Chen and Su Citation2015). ACAT1 (Abdelkreem et al. Citation2019) is involved in ketone body synthesis and degradation and isoleucine catabolism. Another study found that down regulation of ACAT1 was associated with muscle atrophy after spinal cord injury (Huang et al. Citation2021). EHHADH is also related to fatty acid metabolism. HADHA is a subunit of HADH that plays a role in fatty acid β-oxidation (Houten et al. Citation2012). HMGCL is the main enzyme for ketone body synthesis (Puisac et al. Citation2012).
The present study may indicate that the several hub genes screened from DEGs in the amino acid metabolism pathway are related to fatty acid metabolism, indicating that there is an interaction between fatty acid metabolism and amino acid metabolism to regulate the meat quality of grass carp, but the mechanism is still unclear and needs further research.
Conclusions
The outcomes of the current investigation revealed that dietary faba bean increased hardness, shear force and muscle fibre diameter in grass carp. These outcomes indicate that the DEGs of crisp grass carp were concentrated in material metabolism, energy metabolism and ribosome function, and the comprehensive regulation of multiple genes and multiple pathways changes the meat quality of crisp grass carp finally. The interaction between fatty acid metabolism and amino acid metabolism still needs further study.
Author contributions
Meilin Hao and Chuntao Li participated in the design of the study. Wenjie Cheng and Yuxiao Xie did the experiments and did the data analysis. Meilin Hao, Sumei Zhao and Wenjie Cheng drafted the manuscript and all authors contributed to finalising the writing.
Ethical approval
The experiment was carried out in accordance with the research plan of the Institutional Animal Care and Use Committee of Zunyi Normal College (protocol number: ZUNSHIFA[2018]08).
Consent form
All authors agree to participate in the publication of this article.
Supplemental Material
Download MS Excel (7.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the fundings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Abdelkreem E, Harijan RK, Yamaguchi S, Wierenga RK, Fukao T. 2019. Mutation update on ACAT1 variants associated with mitochondrial acetoacetyl-CoA thiolase (T2) deficiency. Hum Mutat. 40(10):1641–1663. doi: 10.1002/humu.23831.
- Adamidou S, Nengas I, Alexis M, Foundoulaki E, Nikolopoulou D, Campbell P, Karacostas I, Rigos G, Bell GJ, Jauncey K. 2009. Apparent nutrient digestibility and gastrointestinal evacuation time in European seabass (Dicentrarchus labrax) fed diets containing different levels of legumes. Aquaculture. 289(1–2):106–112. doi: 10.1016/j.aquaculture.2009.01.015.
- Agarwal M, Sharma A, Kumar P, Kumar A, Bharadwaj A, Saini M, Kardon G, Mathew SJ. 2020. Myosin heavy chain-embryonic regulates skeletal muscle differentiation during mammalian development. Development. 147(7): dev184507. doi: 10.1242/dev.184507.
- Azaman SNA, Satharasinghe DA, Tan SW, Nagao N, Yusoff FM, Yeap SK. 2019. Identification and analysis of microRNAs in Chlorella sorokiniana using high-throughput sequencing. Genes. 11(10): 1131. doi: 10.3390/genes11101131.
- Buszczak M. 2023. Ribosome homeostasis. Semin Cell Dev Biol. 136:1–2. doi: 10.1016/j.semcdb.2022.07.008.
- Canosa LF, Bertucci JI. 2020. Nutrient regulation of somatic growth in teleost fish. The interaction between somatic growth, feeding and metabolism. Mol Cell Endocrinol. 518:111029. doi: 10.1016/j.mce.2020.111029.
- Chen Y, Su Z. 2015. Reveal genes functionally associated with ACADS by a network study. Gene. 569(2):294–302. doi: 10.1016/j.gene.2015.05.069.
- Chen YY, Zhang XN, Xu CZ, Zhou DH, Chen J, Liu ZX, Sun Y, Huang W, Qu LS. 2021. MCCC2 promotes HCC development by supporting leucine oncogenic function. Cancer Cell Int. 21(1):22. doi: 10.1186/S12935-020-01722-W.
- de Carvalho CCCR, Caramujo MJ. 2018. The various roles of fatty acids. Molecules. 23(10):2583. doi: 10.3390/molecules23102583.
- Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. 2018. Lactate dehydrogenase A: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 7(12):6124–6136. doi: 10.1002/cam4.1820.
- Fry CS, Kirby TJ, Kosmac K, McCarthy JJ, Peterson CA. 2017. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell. 20(1):56–69. doi: 10.1016/j.stem.2016.09.010.
- Fu S, Wang B, Zhu Y, Xue Y, Zhong W, Miao Y, Du Y, Wang A, Wang L. 2022. Effects of faba bean (Vicia faba) diet on amino acid and fatty acid composition, flesh quality and expression of muscle quality-related genes in muscle of “crispy” grass carp, Ctenopharyngodon idella. Aquacult Res. 53(13):4653–4662. doi: 10.1111/are.15957.
- Genuth NR, Barna M. 2018. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol Cell. 71(3):364–374. doi: 10.1016/j.molcel.2018.07.018.
- Gill N, Dhillon B. 2022. RNA-seq data analysis for differential expression. Methods in molecular biology (Clifton, NJ), 2391: 45–54. doi: 10.1007/978-1-0716-1795-3_4.
- Guetg A, Mariotta L, Bock L, Herzog B, Fingerhut R, Camargo SMR, Verrey F. 2015. Essential amino acid transporter Lat4 (Slc43a2) is required for mouse development. J Physiol. 593(5):1273–1289. doi: 10.1113/jphysiol.2014.283960.
- Houten SM, Denis S, Argmann CA, Jia Y, Ferdinandusse S, Reddy JK, Wanders RJA. 2012. Peroxisomal l-bifunctional enzyme (Ehhadh) is essential for the production of medium-chain dicarboxylic acids. J Lipid Res. 53(7):1296–1303. doi: 10.1194/jlr.M024463.
- Hu L, Ren S, Shen Q, Ye X, Chen J, Ling J. 2018. Protein oxidation and proteolysis during roasting and in vitro digestion of fish (Acipenser gueldenstaedtii). J Sci Food Agric. 98(14):5344–5351. doi: 10.1002/jsfa.9075.
- Huang H, Xue J, Zheng J, Tian H, Fang Y, Wang W, Wang G, Hou D, Lin J. 2021. Bioinformatic analysis of the gene expression profile in muscle atrophy after spinal cord injury. Sci Rep. 11(1):21903. doi: 10.1038/s41598-021-01302-6.
- Huo W, Weng K, Gu T, Luo X, Zhang Y, Zhang Y, Xu Q, Chen G. 2021. Effects of integrated rice-duck farming system on duck carcass traits, meat quality, amino acid, and fatty acid composition. Poult Sci. 100(6):101107. doi: 10.1016/J.PSJ.2021.101107.
- Jiang W, Li X, Xu X, Ma M, Leng X. 2020. The supplementation of nutrient additives in broad bean-based diet improved the growth of “crisped” grass carp, Ctenopharyngodon idellus. J World Aquacult Soc. 51(1):299–311. doi: 10.1111/jwas.12637.
- Johansen KA, Overturf K. 2006. Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 144(1):119–127. doi: 10.1016/j.cbpb.2006.02.001.
- Knowles TG, Brown SN, Warriss PD, Lines J, Tinarwo A, Bravo A, Carvalho H, Gonçalves A. 2007. Effect of electrical stunning at slaughter on the carcass, flesh and eating quality of farmed sea bass (Dicentrarchus labrax). Aquacult Res. 38:1732–1741. doi: 10.1111/j.1365-2109.2007.01846.x.
- Li Q, Huang Y, Zhang X, Zou C, Lin L. 2023. Improvement of muscle quality in tilapia (Oreochromis niloticus) with dietary faba bean (Vicia faba L.). Front Nutr. 10:1153323. doi: 10.3389/FNUT.2023.1153323.
- Li S, Liu H, Bai J, Zhu X. 2017. Transcriptome assembly and identification of genes and SNPs associated with growth traits in largemouth bass (Micropterus salmoides). Genetica. 145(2):175–187. doi: 10.1007/s10709-017-9956-z.
- Li X, Chen S, Sun J, Huang X, Tang H, He Y, Pan Q, Gan L. 2020. Partial substitution of soybean meal with faba bean meal in grass carp (Ctenopharyngodon idella) diets, and the effects on muscle fatty acid composition, flesh quality, and expression of myogenic regulatory factors. J World Aquacult Soc. 51(5):1145–1160. doi: 10.1111/jwas.12671.
- Lin W-L, Zeng Q-X, Zhu Z-W. 2009. Different changes in mastication between crisp grass carp (Ctenopharyngodon idellus C. et V) and grass carp (Ctenopharyngodon idellus) after heating: the relationship between texture and ultrastructure in muscle tissue. Food Res Int. 42(2):271–278. doi: 10.1016/j.foodres.2008.11.005.
- Liu Q, Xu B, Xiao T, Su J, Zhong L. 2013. Molecular cloning, characterization and expression analysis of coagulation factor VII gene in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 35(2):618–622. doi: 10.1016/j.fsi.2013.05.015.
- Lu X, Chen H-M, Qian X-Q, Gui J-F. 2020. Transcriptome analysis of grass carp (Ctenopharyngodon idella) between fast- and slow-growing fish. Comp Biochem Physiol Part D Genomics Proteomics. 35(2020):100688. doi: 10.1016/j.cbd.2020.100688.
- Ma L-L, Kaneko G, Wang X-J, Xie J, Tian J-J, Zhang K, Wang G-J, Yu D-G, Li Z-F, Gong W-B, et al. 2020. Effects of four faba bean extracts on growth parameters, textural quality, oxidative responses, and gut characteristics in grass carp. Aquaculture. 516:734620. doi: 10.1016/j.aquaculture.2019.734620.
- Madureira TV, Castro LFC, Rocha E. 2016. Acyl-coenzyme A oxidases 1 and 3 in brown trout (Salmo trutta f. fario): can peroxisomal fatty acid β-oxidation be regulated by estrogen signaling? Fish Physiol Biochem. 42(1):389–401. doi: 10.1007/s10695-015-0146-6.
- Puisac B, Ramos M, Arnedo M, Menao S, Gil-Rodríguez MC, Teresa-Rodrigo ME, Pié A, de Karam JC, Wesselink J-J, Giménez I, et al. 2012. Characterization of splice variants of the genes encoding human mitochondrial HMG-CoA lyase and HMG-CoA synthase, the main enzymes of the ketogenesis pathway. Mol Biol Rep. 39(4):4777–4785. doi: 10.1007/s11033-011-1270-8.
- Qu Y-Y, Zhao R, Zhang H-L, Zhou Q, Xu F-J, Zhang X, Xu W-H, Shao N, Zhou S-X, Dai B, et al. 2020. Inactivation of the AMPK–GATA3–ECHS1 pathway induces fatty acid synthesis that promotes clear cell renal cell carcinoma growth. Cancer Res. 80(2):319–333. doi: 10.1158/0008-5472.CAN-19-1023.
- Rao Y, Su J. 2015. Insights into the antiviral immunity against grass carp (Ctenopharyngodon idella) reovirus (GCRV) in grass carp. J Immunol Res. 2015:670437. doi: 10.1155/2015/670437.
- San J, Du Y, Wu G, Xu R, Yang J, Hu J. 2021. Transcriptome analysis identifies signaling pathways related to meat quality in broiler chickens – the extracellular matrix (ECM) receptor interaction signaling pathway. Poult Sci. 100(6):101135. doi: 10.1016/J.PSJ.2021.101135.
- Tian J-J, Ji H, Wang Y-F, Xie J, Wang G-J, Li Z-F, Yu E-M, Yu D-G, Zhang K, Gong W-B. 2019. Lipid accumulation in grass carp (Ctenopharyngodon idellus) fed faba beans (Vicia faba L.). Fish Physiol Biochem. 45(2):631–642. doi: 10.1007/s10695-018-0589-7.
- von Walden F. 2019. Ribosome biogenesis in skeletal muscle: coordination of transcription and translation. J Appl Physiol (1985). 127(2):591–598. doi: 10.1152/japplphysiol.00963.2018.
- Wu C-S, Ma Z-Y, Zheng G-D, Zou S-M, Zhang X-J, Zhang Y-A. 2022. Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution. BMC Genomics. 23(1):271. doi: 10.1186/s12864-022-08503-x.
- Wu G. 2013. Functional amino acids in nutrition and health. Amino Acids. 45(3):407–411. doi: 10.1007/s00726-013-1500-6.
- Xu W-H, Guo H-H, Chen S-J, Wang Y-Z, Lin Z-H, Huang X-D, Tang H-J, He Y-H, Sun J-J, Gan L. 2020. Transcriptome analysis revealed changes of multiple genes involved in muscle hardness in grass carp (Ctenopharyngodon idellus) fed with faba bean meal. Food Chem. 314:126205. doi: 10.1016/j.foodchem.2020.126205.
- Yang G, Zhao W, Qin C, Yang L, Meng X, Lu R, Yan X, Cao X, Zhang Y, Nie G. 2020. Igfbp3 in grass carp (Ctenopharyngodon idellus): molecular identification and mRNA expression under glucose, insulin and glucagon. Comp Biochem Physiol B Biochem Mol Biol. 242:110394. doi: 10.1016/j.cbpb.2019.110394.
- Yang H, Xu Z, Xu X, Mizanur Rahman M, Li X, Leng X. 2022. Transcriptomic and biochemical analyses revealed the improved growth, lipid metabolism and flesh quality of grass carp (Ctenopharyngodon idellus) by dietary Eucommia ulmoides bark and leaf supplementation. J Anim Sci. 100(10):1–11. doi: 10.1093/JAS/SKAC250.
- Yang S, Li L, Qi B, Wu Y, Hu X, Lin W, Hao S, Huang H. 2015. Quality evaluation of crisp grass carp (Ctenopharyngodon idellus C. et V) based on instrumental texture analysis and cluster analysis. Food Anal Methods. 8(8):2107–2114. doi: 10.1007/s12161-015-0101-2.
- Yu E, Fu B, Wang G, Li Z, Ye D, Jiang Y, Ji H, Wang X, Yu D, Ehsan H, et al. 2020. Proteomic and metabolomic basis for improved textural quality in crisp grass carp (Ctenopharyngodon idellus C.et V) fed with a natural dietary pro-oxidant. Food Chem. 325:126906. doi: 10.1016/j.foodchem.2020.126906.
- Yu E, Xie J, Wang G, Yu D, Gong W, Li Z, Wang H, Xia Y, Wei N. 2014. Gene expression profiling of grass carp (Ctenopharyngodon idellus) and crisp grass carp. Int J Genomics. 2014:639687. doi: 10.1155/2014/639687.
- Yu E-M, Ma L-L, Ji H, Li Z-F, Wang G-J, Xie J, Yu D-G, Kaneko G, Tian J-J, Zhang K, et al. 2019. Smad4-dependent regulation of type I collagen expression in the muscle of grass carp fed with faba bean. Gene. 685:32–41. doi: 10.1016/j.gene.2018.10.074.
- Yu E-M, Zhang H-F, Li Z-F, Wang G-J, Wu H-K, Xie J, Yu D-G, Xia Y, Zhang K, Gong W-B. 2017. Proteomic signature of muscle fibre hyperplasia in response to faba bean intake in grass carp. Sci Rep. 7(1):45950. doi: 10.1038/srep45950.
- Zhang S, Zhang J, Cao C, Cai Y, Li Y, Song Y, Bao X, Zhang J. 2022. Effects of different rearing systems on lueyang black-bone chickens: meat quality, amino acid composition, and breast muscle transcriptome. Genes. 13(10):1898. doi: 10.3390/GENES13101898.
- Zhao X, Wang C, Wang Y, Zhou L, Hu H, Bai L, Wang J. 2020. Weighted gene co-expression network analysis reveals potential candidate genes affecting drip loss in pork. Anim Genet. 51(6):855–865. doi: 10.1111/age.13006.
- Zhou L, Lin K-T, Gan L, Sun J-J, Guo C-J, Liu L, Huang X-d 2019. Intestinal microbiota of grass carp fed faba beans: a comparative study. Microorganisms. 7(10):465. doi: 10.3390/microorganisms7100465.
- Zimmermann MT, Jia K, Jernigan RL. 2016. Ribosome mechanics informs about mechanism. J Mol Biol. 428(5 Pt A):802–810. doi: 10.1016/j.jmb.2015.12.003.