ABSTRACT
Nipah virus (NiV) is a high-risk pathogen which can cause fatal infections in humans. The Indian isolate from the 2018 outbreak in the Kerala state of India showed ~ 4% nucleotide and amino acid difference in comparison to the Bangladesh strains of NiV and the substitutions observed were mostly not present in the region of any functional significance except for the phosphoprotein gene. The differential expression of viral genes was observed following infection in Vero (ATCC® CCL−81™) and BHK−21 cells. Intraperitoneal infection in the 10–12-week-old, Syrian hamster model induced dose dependant multisystemic disease characterized by prominent vascular lesions in lungs, brain, kidney and extra vascular lesions in brain and lungs. Congestion, haemorrhages, inflammatory cell infiltration, thrombosis and rarely endothelial syncitial cell formation were seen in the blood vessels. Intranasal infection resulted in respiratory tract infection characterised by pneumonia. The model showed disease characteristics resembling the human NiV infection except that of myocarditis similar to that reported by NiV-Malaysia and NiV-Bangladesh isolates in hamster model. The variation observed in the genome of the Indian isolate at the amino acid levels should be explored further for any functional significance.
Introduction
Nipah virus (NiV) was first discovered during an encephalitis outbreak in the state of Perak, Malaysia in 1998 in pig farmers [Citation1,Citation2]. The disease was later reported from other states of Malaysia and countries like Singapore, Bangladesh, Philippines and India [Citation1,Citation3–6]. The virus belongs to the genus Henipavirus of family Paramyxoviridae. In India, the NiV outbreak has been reported from two geographically distant regions i.e., from the West Bengal state bordering Bangladesh and Kerala state which is in the southern part of the country [Citation7–9]. The outbreaks reported from Silguri (2001), Nadia (2007) and Kozhikode, Kerala (2018), in India have shown very high fatality rate [Citation4,Citation7,Citation9].
The NiV genome encodes for six structural proteins i.e., the nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), glycoprotein (G) and polymerase (L); and three accessory proteins i.e., C, V and W. The genomic studies have shown the NiV strains identified from the Malaysia (NiV-M) and Bangladesh (NiV-B) shows about ~ 9% nucleotide difference in their genomes suggesting evolution of the virus in the local animal reservoirs in the outbreak areas [Citation10]. Genetic analysis of the NiV sequences from India i.e. the West Bengal and Kerala states showed it to be sub clustering within the NiV-B clade. The later was proposed to be a separate genotype as “I” [Citation9,Citation11]. Many amino acid substitutions were observed in the viral structural and non-structural proteins especially the P gene products of NiV-B isolates which are linked to the regulation of host immune response and virulence [Citation12,Citation13].
The phenotypic characteristics of NiV-M and NiV-B strains also differed in terms of incubation period, fatality rate, transmission pattern and symptoms [Citation1,Citation2,Citation14,Citation15]. Systemic vasculitis was reported from NiV human cases in Malaysia whereas such reports are not available from Bangladesh outbreaks [Citation16]. Acute respiratory distress, myocarditis and encephalitis were the reported clinical findings from the Kerala outbreak and the autopsy of a deceased patient revealed lung involvement with vasculitis [Citation17,Citation18]. Multiple animal models have been described for NiV and Golden Syrian hamsters appears to be a promising model [Citation19–21].
The NiV isolate sequences from Bangladesh showed marked heterogeneity [Citation14]. Till date all the pathogenicity studies using the Bangladesh isolates in animal model have used a 2004 NiV-B (Rajbari) isolate [Citation20–22]. Here we have used NiV isolate obtained from a patient during the Kerala outbreak of 2018 from India which caused about 90% case fatality rate for studying the genomic characteristics, transcription pattern and disease characteristics in hamster model.
Materials and methods
Ethics statement
All the animal experiments were performed in the Biosafety Level (BSL) −4 facility of the Indian Council of Medical Research-National Institute of Virology (ICMR-NIV), Pune. The experiments were approved by the Institutional Animal Ethics committee (Approval no. IAEC/2019/MCL/10) and were performed as per the guidelines of Committee for Control and Supervision of Experiments on Animals (CCSEA), Government of India.
Virus and cells
NiV isolated from the throat swab sample of a patient during the 2018 outbreak in Kerala state, India was used [Citation11]. The isolate (Accession no.: MH523642.1) was passaged four times in Vero (ATCC® CCL−81™) cells (ATCC, USA) and the titre of the virus was 1.4 × 106 median Tissue Culture Infectious Dose (TCID50)/millilitre (ml). The isolate was further sequence verified using Mega 7.2 software.
To understand the NiV growth kinetics, Vero (ATCC® CCL−81™) cells (ATCC, USA) in confluent six well plates were infected with 0.1 mutiplicity of infection (MOI). The plates were incubated for an hour at 37°C in a CO2 incubator. After one hour, the cells were washed with 1× PBS and were incubated with 2% foetal bovine serum supplemented minimal essential media (MEM) at 37°C. The cell culture supernatant was collected at 24-hour intervals and were titrated in Vero (ATCC® CCL−81™) cells and titres were determined by the Reed and Muench method.
Transcriptome analysis
Vero (ATCC® CCL−81™) cells and baby hamster kidney cells (BHK−21) seeded in 6-well tissue culture plates were infected with NiV (1.4 × 105 TCID50/100 µL, 0.1 MOI). Supernatants and cells were harvested at 1 hour (h), 24 h, 40 h, 48 h, and 72 h after infection. Supernatants treated with RNase A (10 µg/ml) for 1 h at 37°C were used as a control. Total RNA was extracted using the Tripure method followed by column purification with a Qiagen viral RNA extraction kit. Poly-A-containing mRNAs were purified from total RNA using oligo-dT beads provided in the Illumina TruSeq Stranded mRNA LT Sample Preparation Kit. cDNA libraries were prepared and sequenced using the Illumina MiniSeq platform using the mid-output cartridge (300 cycles, 150 × 2 paired-end sequencings).
Total reads from both the cell lines were mapped with the reference genome (accession number: AY988601). Transcription of each gene was quantified using the reads per kilobase million (RPKM) method for each time point [Citation23]. Read coverage and RPKM values were determined using CLC Genomics Workbench Software (version 11, Qiagen Bioinformatics).
Lethal dose 50 (LD50) determination
For the experiments, female Golden Syrian hamsters (Mesocricetus auratus) of 10–12-week-old age procured from a CCSEA approved facility were used. The animals were housed in individually ventilated cages in the containment facility with ad libitum access to food and water. The hamsters were randomly assigned to fourteen experimental groups (n = 5 hamsters/group) after a week period of acclimatization in the facility. Six virus dilutions ranging from 1.4 × 106 to 1.4 × 10° TCID50/ml dose were used for LD50 determination. For the intraperitoneal LD50, five hamsters each were infected with 0.5 ml of virus doses through intraperitoneal route under isoflurane anaesthesia. A group of five hamsters were kept as uninfected control. For the LD50 assessment by intranasal route, 0.1 ml of NiV was used with same virus dilutions as used for the intraperitoneal and one group was kept as control after mock infection with 0.1 ml sterile media. The animals were monitored twice daily for a period of 28 days. Lethal dose 50 was calculated by Reed and Muench method considering death and more than 15% body weight loss as endpoints. The percent survival was plotted for the intraperitoneal and intranasal LD50 studies with the number of animals which survived the infection with < 15% or without weight loss.
Pathogenicity experiments
Thirty Syrian hamsters of 10–12 weeks of age were used. Twenty-five hamsters were infected with 7 × 105 TCID50 virus dose (100 × LD50 dose) intraperitoneally and four hamsters each were sequentially sampled and euthanized on day 1,3,5,7, and 9 days post infection. Blood, throat swab, nasal wash, conjunctival swab, and rectal swab were collected from animals before euthanasia. All the visceral organs and brain samples were collected after necropsy for viral RNA/viral load estimation, cytokine estimation as well as for histopathology. For intranasal pathogenicity assessment, fifteen Syrian hamsters were infected intranasally with a dose of 1.4 × 105 TCID50. Four animals each were euthanized on day 3, 5, and 7 to collect the throat swab, nasal wash, rectal swab, blood and organs for viral RNA load and histopathology. Five animals were kept as uninfected control for both the experiments.
Real time quantitative PCR (RT-qPCR)
The samples (serum/swab/tissue homogenate) in lysis buffer were transferred to the BSL−2 facility for RNA extraction. The extraction was performed using Magmax Viral RNA isolation kit (Thermoscientific, USA) and RT-qPCR was performed using published primers for NiV N gene as described earlier [Citation24]. For cytokine and chemokine mRNA (IL−1, IL−4, IL−6, IL−10, IL−12 and IFN-γ) quantification, published primers were used [Citation25]. The RNA concentrations of the samples were adjusted as that of the uninfected control animal samples and relative quantification were performed. HPRT gene was used as an internal control. The fold change was calculated based on delta-delta Ct method.
Anti-NiV hamster immunoglobulin G ELISA
The detection was performed using an in-house developed enzyme linked immunosorbent assay (ELISA) using gamma irradiated (24 KGy irradiation dose) Nipah virus whole antigen. Four rows of the ELISA plates were coated with inactivated NiV antigen and four rows with normal Vero (ATCC® CCL−81™) cell lysate as negative antigen. After overnight coating at 4°C, liquid plate sealer (Candor, USA) was used to block the wells. After washing the plates, 100 µl of hamster serum samples were added to both positive and negative antigen wells in 1: 100 dilutions. The plates were incubated at 37°C for one hour followed by washing using the wash buffer. Anti-hamster IgG Horseradish Peroxidase antibodies (1:3000 dilution) were added and incubated for 60 minutes at 37°C. 3,3′,5,5′-Tetramethylbenzidine substrate was added and incubated for 10 minutes for colour development and the reaction was terminated and plates were read at 450 nm.
Virus titration
Tenfold dilution of the samples was added to Vero (ATCC® CCL−81™) cell monolayers and incubated for 1 hour at 37°C. After removing the inoculum, the media containing 2× carboxymethyl cellulose in MEM was used and incubated for 5 days. The media was decanted and the plates were counted for plaques. The titre was determined by Reed and Muench method and was expressed as TCID50/ml.
Histopathology
The organ samples were fixed in 10% neutral buffered formalin and were processed by routine techniques for Haematoxylin and Eosin staining as described earlier [Citation26]. The slides were blindly screened and evaluated by two pathologists.
Results
Genome characterization
The nucleotide and amino acid identity of the Indian isolate was about 97% and 95% with the NiV-B sequences whereas it was about 91% and 83% with the NiV-M sequences respectively (). The amino acid substitutions in the structural and non-structural proteins of the isolate in comparison to the NiV-B and NiV-M isolates are given in the supplementary table S1. The substitutions in the N protein except the 387N were not in the minimum contiguous sequence (30–404 position) and in the four conserved hydrophobic regions demonstrated for the capsid assembly [Citation27]. The reported P binding regions (1-54,135–146, 468–496) of the N protein were found conserved in the Indian isolate [Citation28–30]. The substitutions described in the signal transducer and activator of transcription (STAT)−1 binding domain (position 114–140) of the P/V/W protein known to decrease the STAT−1 binding were not present in the Indian isolate [Citation31–33]. The first 38 residues of P important for N binding were conserved in the isolate whereas C terminal domain which binds to the N protein showed many substitutions in the NiV-B and Indian isolate [Citation28,Citation34]. Numerous substitutions were found between the 100–130 position of P/V/W protein important for STAT−2 binding whereas the motifs between the residues 230 and 237 which are critical were found conserved [Citation35]. The other substitutions observed in the P/W/V proteins were not located in any positions described for the STAT binding [Citation4,Citation28,Citation31,Citation32]. A single substitution was observed within residues 81 to 113 important for polymerase cofactor function of P at 105 position in the Indian isolate whereas the critical residue R555 defined was intact [Citation32,Citation36]. The N and C terminal residues of the C protein were found conserved except at the position Y98H. Apart from the NiV-B sequences, I40T and Y98H were found in the C protein of the Indian isolate. The amino acid motifs YMYL and YPLGVG important for budding in the M protein were found intact in the isolate [Citation37,Citation38]. No mutations were found in the amino acid residues in the globular head of G protein which can affect the fusion promotion capacity and ephrin binding [Citation39,Citation40] and in the F protein cytoplasmic tail motifs important for fusion and protein trafficking [Citation41]. The predicted mutations in the NiV glycoprotein which can affect the binding of m102.4 monoclonal antibody were absent in the Indian isolate [Citation42]. The substitutions found in the L protein were same as that of the NiV-B sequences.
Table 1. The percent nucleotide and amino acid identity of the NiV isolates with NiV-M reference isolate (NC002728).
Replication kinetics and transcriptome analysis
The growth curve in Vero (ATCC® CCL−81™) cells showed an exponential increase in TCID50 from day 1 to reach peak titre by day 3 followed by decrease in the further days (. The cytopathic effects (cell rounding, aggregation, syncitia formation) were observed in Vero (ATCC® CCL−81™) cells from day 1 post infection (). The time point study analysis showed high expression levels for M,P/V/W/C,N mRNAs in the infected Vero (ATCC® CCL−81™) cells and intermediate levels for F and lower levels for G and L (). The CPE were observed in the BHK−21 cells from 3 days post infection (). The transcriptional pattern in BHK−21 cells showed higher expression levels of M and P/V/W/C mRNAs, intermediate levels for the N, F and G; and lower levels of L ().
Figure 1. Characterization of NiV in cells: Growth kinetics in the Vero (ATCC® CCL − 81™) cells expressed in a) TCID50 and b) viral RNA copy number/ml. c) Vero CCL − 81 cells showing cytopathic effects after 1 day post NiV infection, and d)cell control. Differential expression of NiV genes in e) Vero-CCL81 cells expressed as RPKM values at different time points. f) BHK − 21 cells infected with NiV showing cytopathic effect on 3 days post infection and g) BHK − 21 cell control. Differential expression of NiV genes in h) BHK − 21 cells expressed as RPKM values at different time points.
Lethal dose 50
For the intraperitoneal route of infection, LD50 was found to be 7000 TCID50 (). Sickness was observed in virus dilutions ranging from 7 × 105 TCID50 to 7 × 102 TCID50 dose. The signs observed were ruffled hair coat, slow movement, head tilting, circling, hind limb paralysis, blood-tinged nasal discharge, and body weight loss (). The disease onset in hamsters infected with 7 × 105 TCID50 was faster (within 5 to 7 days) compared to the further virus dilutions. In hamsters inoculated with 7 × 104, 7 × 103 and 7 × 102 TCID50, disease onset was by 7 to 11, 9 to 15 and 9 to 15 days respectively. No overt clinical signs or seroconversion were observed in hamsters infected with further lower dilutions.
Figure 2. Survival and body weight changes in hamsters after NiV infection. a) Percent survival and b) body weight change in hamsters post intraperitoneal infection. c) Percent survival and d) body weight change in hamsters post intranasal infection.
In case of intranasal route of infection, limited body weight gain was observed and lethality was observed only in 1/5 animals of 1.4 × 105 on day 12 and in 1.4 × 104 TCID50 inoculated animals on day 18 (). Seroconversion was observed in 1.4 × 105 and 1.4 × 104 TCID50 infected groups.
Pathogenicity by intraperitoneal route of infection
Nasal wash showed the presence of viral RNA till day 7 with peak on day 5 in most of the hamsters and in ¼ hamsters on day 9. Conjunctival swabs of only two severely sick hamsters on day 5 and 1 sick hamster on day 9 showed viral RNA positivity. The rectal swab, oral swab and nasal wash samples of these three hamsters showed viral RNA positivity. Other than this, none of the animals showed oral or conjunctival RNA positivity. Rectal swab of two hamsters showed viral RNA positivity on day 1 (). Viremia was observed from day 1 [mean genome copies/ml ± standard deviation (SD) = 5955.8 ± 5401.8] till day 7 (1290 ± 2581.3) with a peak on day 3 (20345 ± 13528) ().
Figure 3. NiV RNA shedding and viremia in hamsters post intraperitoneal infection. a) Viral shed through the nasal wash, oral and rectal route. b) Viral RNA in serum samples collected from hamsters on various time points. The individual values along with median value are plotted in the graph.
The organ viral RNA load observed was highest in brain followed by lungs, kidney, liver, heart, trachea, spleen and intestine (). In brain, kidney and heart, viral RNA load was found increasing, reaching a maximum by day 9. In lungs, peak viral RNA load was seen on day 5 (mean genome copies/ml ± SD = 5.0 × 107 ±45395035.73) which decreased on further time points. On virus titration, brain showed highest virus titre.
Figure 4. NiV load in organs of hamsters after intraperitoneal infection. Viral RNA load in a) brain b) heart c) lungs d) trachea e) intestine f) spleen g) liver and h) kidney. i) Live virus load in brain, lungs, spleen and kidney. The individual values along with median value are plotted in the graph.
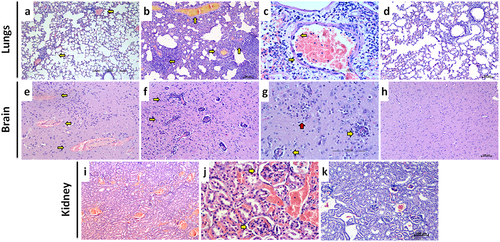
The major histopathological changes observed in the organs were thrombosis, vasculitis in brain and lungs, severe engorgement of the blood vessels of kidney and liver (). In lungs, congestion of the large vessels as well as alveolar capillaries, haemorrhages in the parenchyma, thrombus formation and vasculitis in the vessels were observed. Vasculitis in lungs was evident from day 3 post infection and inflammatory changes were observed in the nearby alveolar parenchyma along with haemorrhages. Degenerative changes in the bronchiolar epithelium, oedema, alveolar septal thickening and haemorrhages in parenchyma became prominent from 5th day post infection. Two hamsters each which were severely sick on day 4 and 6 post infection showed diffuse haemorrhages in lungs. Few blood vessels in the lungs also showed endothelial cell syncitia formation. In the kidney, severe congestive changes in the renal cortex were observed from 1D day post infection. The glomerular and peritubular capillaries showed severe engorgement and thrombosis was evident in the glomerular capillaries in the further days. In brain, perivascular cuffing, gliosis and degenerative changes were observed in two hamsters euthanized on day 7 and 4 hamsters euthanized on day 9. Congestive changes and haemorrhages were observed in the ependymal blood vessels. Rarely, haemorrhages were observed in the brain and perivascular space. Liver parenchyma also showed diffuse vascular and sinusoidal engorgement.
Figure 5. Histopathological observations in NiV infected hamsters after intraperitoneal infection. a) Lung section showing multifocal perivascular inflammatory cell infiltration in the alveolar parenchyma (arrows) on 3 days post infection, (H& E, scale bar = 100 µm). b) Lung section showing vasculitis, thrombosis and diffuse alveolar thickening on 7 days post infection, (H& E, scale bar = 100 µm) c) Blood vessel in the lungs showing multiple endothelial cell syncitia formation (arrows) and perivascular mononuclear cell infiltration, (H& E, scale bar = 200 µm) d) Lung section showing normal histological features, (H& E, scale bar = 200 µm) e) Brain section showing blood vessels with haemorrhage in the perivascular space, (H& E, scale bar = 200 µm). f) Brain section showing multiple blood vessels with perivascular cuffing, (H& E, scale bar = 100 µm). g) Brain section showing blood vessels with perivascular cuffing (yellow arrows) and gliosis (red arrow), (H& E, scale bar = 200 µm). h) Brain section showing normal histological features, (H& E, scale bar = 100 µm) i) Kidney section showing diffuse vascular engorgement, (H& E, scale bar = 200 µm).J) Kidney section showing congested blood vessels and thrombotic plugs in the glomerular capillaries, (H& E, scale bar = 200 µm).K) Kidney section showing normal histological features, (H& E, scale bar = 100 µm).
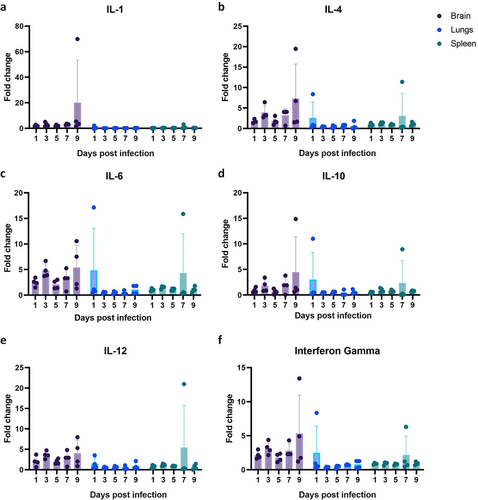
Four hamsters which survived were observed till 28 days and the anti-NiV IgG response was observed from day 18 in these hamsters. The cytokine levels i.e. IL−4, IL−6, IL−12 and IFN-Gamma in brain samples were found slightly upregulated compared to the lungs and spleen samples at various time points (). A mean 5-fold increase was seen in the brain samples. IL−4, IL−6, IL−10, and IFN-Gamma were highest at the 9 days post infection. The spleen and lung samples did not show any consistent upregulation of the host genes studied.
Pathogenicity by intranasal infection in hamsters
No respiratory or neurological signs were observed in animals post infection. Viral RNA shedding was observed through the oral, nasal and alimentary tract in these infected animals (). In contrast to the intraperitoneal infection, we could not observe any viremia and viral RNA in brain in animals by intranasal infection (). Lungs showed the maximum viral RNA load followed by liver, kidney and spleen. Viral RNA could be detected in heart of only one animal each on day 3 and 7. Intestine showed viral positivity till day 5. Lungs sections showed severe broncho-interstitial pneumonia, vasculitis and thrombosis from 3 days after infection in these animals. Severe congestion and thrombosis were evident in the glomerular capillaries. All the other organs including the brain showed normal histology.
Figure 7. Viral RNA load in hamsters post intranasal NiV infection. A) Viral RNA shed through the nasal wash, oral and rectal route. Viral RNA in B) lungs c) liver d) heart e) intestine f) kidney and spleen samples collected from hamsters on various time points. The individual values along with median value are plotted in the graph.
Discussion
In the present study, we have characterized the NiV isolate obtained from a patient during the outbreak in Kerala, India. The nucleotide and amino acid identity observed in the Indian and NiV-B strains with the NiV-M were similar with around < 0.5% difference. The majority of substitutions observed in the Indian isolate viral proteins were similar to that already reported for NiV-B genotype. These changes are mostly not present in the region of any functional significance which is defined for NiV except for the P gene, the importance of which needs to be studied further. The growth kinetics observed were similar to that reported for NiV-M in Vero cells [Citation43]. BHK−21 cells were permissive for infection as demonstrated earlier [Citation20]. In the mRNA expression study, M gene mRNAs were the most abundant in both the cells, followed by the P/V/W/C and N. Non-segmented ssRNA viruses differentially express their genes, with the level of transcripts regulated by their position relative to the single promoter [Citation44].
The isolate was found pathogenic in hamsters and we observed two types of diseases, i.e., rapid onset respiratory disease and a delayed onset nervous sign [Citation20,Citation45,Citation46]. Respiratory/neurological signs and lethality by both intranasal and intraperitoneal routes have been demonstrated with the NiV-B in hamster model [Citation20]. The signs in hamsters can range from mild (hypo activity or abnormal gait) to severe (severe dyspnoea, neurological signs, weight loss, death) and animal may recover completely[Citation45,Citation46] . Different combinations of acute respiratory distress syndrome (ARDS), encephalitis and myocarditis were reported in the NiV disease in humans [Citation17].
The virus dose, strain and route of infection plays role in pathogenicity in hamsters [Citation47]. Wong et al reported a 174 fold higher LD50 dose for NiV-M intranasal infection in comparison to the intraperitoneal infection [Citation47]. This could probably due to the faster access of the virus to the systemic circulation after the intraperitoneal infection. The hamsters infected with the lower doses like 10 or 100 pfu did not show any clinical signs or seroconversion in the above study as we observed here [Citation47]. Intranasal infection showed only limited virus spread probably due to the lesser dose used in the present study for the intranasal infection and the multiple immunological barriers which need to be encountered for successful viral replication and spread. A faster disease progression was observed with NiV-M strain (with seven times higher LD50) compared to NiV-B strain in hamsters [Citation20]. The variation in the mortality observed with different administration routes could be due to the difference in the virus doses used.
The intranasal route of infection may be more appropriate for transmission studies using hamster model as it showed consistent viral RNA shedding. Even though viral RNA could be detected in the nasal cavity till day 7 in the intranasally infected hamsters, the presence of viral RNA could not be seen in the brain of these animals. Viremia was also not observed in intranasally infected hamsters. This indicates that blood cell mediated entry could be a more probable route for CNS infection rather than the cranial nerves [Citation48,Citation49]. Olfactory epithelial infection and transport to the brain have been reported in hamsters [Citation50].
Viremia, vascular changes observed in multiple organs and endothelial cell syncitia formation are in agreement with the endothelial tropism of the virus due to the ephrin B2/B3 receptor expression [Citation51,Citation52]. Tropism of NiV to arterial/arteriolar endothelium has been demonstrated in NiV infected hamsters [Citation53]. The organ changes observed in the present study were similar to as reported for the earlier NiV isolates [Citation19,Citation20,Citation47]. The observations from earlier studies also suggest that differences between the NiV-M and NiV-B outbreaks may not be due to the intrinsic differences in the virus [Citation53]. Cytokine and chemokine mRNAs upregulation observed was not marked similar to the earlier observations of NiV-B infected hamster model [Citation20,Citation46]. Dose dependant cytokine response is demonstrated in NiV infected hamsters [Citation46]. The IL−4 and IL−6 are important for the lymphocyte differentiation and activation [Citation54,Citation55]. Inflammatory cellular infiltration was seen in the hamster brain tissues with prominent perivascular infiltrations. Cytokine upregulation can regulate the blood brain barrier transport. The disruption of blood brain barrier and lymphocyte mediated transinfection has been reported after NiV infection [Citation46,Citation48]. A delayed upregulation of cytokines ie after 10 days post infection has been reported in the brain of intranasally NiV infected hamsters [Citation46]. IL−6, IL−4, IFN-γ, IL−10, and IL−1β were found downregulated in brain during the early stages of NiV infection [Citation46]. There is limitation of reagents for studying immune response in detail in hamster model. We also could not compare with other NiV isolates like NiV-B or NiV-M in the animal model as we don’t possess these isolates with us.
In the present study we have characterized the NiV isolate from 2018 outbreak of India. The majority of the substitutions observed in the isolate were similar to that observed in the NiV-B isolates except in the P gene, the functional significance of which needs to be explored. The virus replication in different organs and characteristic pathological changes in brain, lungs and kidney could be demonstrated in the hamster model. A dose and route dependant pathogenicity and organ involvement were observed in the hamster model.
Supplemental Material
Download MS Word (105.5 KB)Acknowledgements
This study was supported by Indian Council of Medical Research as an intramural grant to ICMR-National Institute of Virology, Pune. We acknowledge the support received from Prof. Priya Abraham, Director, ICMR-NIV, Pune. The authors acknowledge the support of Dr Dimpal Nyayanit for the next generation sequencing analysis and the technical support received from the laboratory team of Maximum Containment Facility of ICMR-NIV, Pune, i.e., Mr Manoj Kadam and Mr Rajen Lakra for animal experiments, Dr Rajlaxmi Jain, Mrs Savita Patil, Mrs Triparna Majumdar, Mrs Kaumudi Kalele, Ms Jyoti Yemul, Ms Pranita Gawande for sample testing and Mr Yash Joshi for support in sequence analysis. We acknowledge the support of Dr N.Kurkure for the histopathology services and Dr B. Dinesh Kumar, Scientist G, ICMR-National Institute of Nutrition, Hyderabad, Telangana, India for providing the animals required for the experiment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article. The transcriptome data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/sra/?term=, reference number. [SRR24202203, SRR24202204, SRR24202205, SRR24202206, SRR24202207, SRR24202208, SRR24202209, SRR24202210, SRR24202211, SRR24202212].
Supplemental data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2023.2224642
Additional information
Funding
References
- Lam SK, Chua KB 2002. Nipah virus encephalitis outbreak in Malaysia. Clin Infect Dis 34 Suppl 2:S48–13. doi:10.1086/338818
- Chua KB, Goh KJ, Wong KT, et al. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257–1259. 9186 doi:10.1016/S0140-6736(99)04299-3
- Paton NI, Leo YS, Zaki SR, et al. 1999. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354:1253–1256. 9186 doi:10.1016/S0140-6736(99)04379-2
- Chadha MS, Comer JA, Lowe L, et al. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 12:235–240. 2 doi:10.3201/eid1202.051247
- Gurley ES, Montgomery JM, Hossain MJ, et al. 2007. Person-to-Person transmission of nipah virus in a Bangladeshi community. Emerg Infect Dis 13:1031–1037. 7 doi:10.3201/eid1307.061128
- Ching PKG, de Los Reyes VC, Sucaldito MN, et al. 2015. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis 21:328–331. 2 doi:10.3201/eid2102.141433
- Arunkumar G, Chandni R, Mourya DT, et al. Outbreak investigation of nipah virus disease in Kerala, India, 2018. J Infect Dis. 2019;219:1867–1878. doi:10.1093/infdis/jiy612
- Sudeep AB, Yadav PD, Gokhale MD, et al. 2021. Detection of Nipah virus in Pteropus medius in 2019 outbreak from Ernakulam district, Kerala, India. BMC Infect Dis 21:162. 1 doi:10.1186/s12879-021-05865-7
- Arankalle VA, Bandyopadhyay BT, Ramdasi AY, et al. 2011. Genomic characterization of Nipah virus, West Bengal, India. Emerg Infect Dis 17:907–909. 5 doi:10.3201/eid1705.100968
- Rahman MA, Hossain MJ, Sultana S, et al. 2012. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis 12:65–72. 1 doi:10.1089/vbz.2011.0656
- Yadav PD, Shete AM, Kumar GA, et al. 2019. Nipah virus sequences from humans and bats during nipah outbreak, Kerala, India, 2018. Emerg Infect Dis 25:1003–1006. 5 doi:10.3201/eid2505.181076
- Lo MK, Lowe L, Hummel KB, et al. Characterization of Nipah Virus from Outbreaks in Bangladesh, 2008–2010. Emerg Infect Dis J - CDC. 2012 February;18(2):248–255. doi: 10.3201/eid1802.111492
- Rahman MZ, Islam MM, Hossain ME, et al. 2021. Genetic diversity of Nipah virus in Bangladesh. Inter J Infect Dis 102:144–151. doi:10.1016/j.ijid.2020.10.041
- Harcourt BH, Lowe L, Tamin A, et al. 2005. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis 11:1594–1597. 10 doi:10.3201/eid1110.050513
- Hossain MJ, Gurley ES, Montgomery JM, et al. 2008. Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis 46:977–984. 7 doi:10.1086/529147
- Wong KT, Shieh W-J, Kumar S, et al., Nipah virus pathology working group. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol 161:2153–2167. 2002 6 doi:10.1016/S0002-9440(10)64493-8
- Chandni R, Renjith TP, Fazal A, et al. 2020. Clinical manifestations of nipah virus–infected patients who presented to the emergency department during an outbreak in Kerala State in India, May 2018. Clin Infect Dis 71:152–157. 1 doi:10.1093/cid/ciz789
- Rajeevan K, Sathi PP, Prasannan K, et al. 2021. Nipah virus infection: autopsy of a clinical challenge. Indian J Pathol Microbiol 64:621. 3 doi:10.4103/IJPM.IJPM_455_20
- Mire CE, Satterfield BA, Geisbert JB, et al. 2016. Pathogenic differences between Nipah virus Bangladesh and Malaysia Strains in Primates: implications for Antibody Therapy. Sci Rep 6:30916. 1 doi:10.1038/srep30916
- DeBuysscher BL, de Wit E, Munster VJ, et al. 2013. Comparison of the pathogenicity of Nipah virus isolates from Bangladesh and Malaysia in the Syrian hamster. PLoS Negl Trop Dis 7:e2024. 1 doi:10.1371/journal.pntd.0002024
- Clayton BA, Middleton D, Bergfeld J, et al. 2012. Transmission routes for Nipah Virus from Malaysia and Bangladesh. Emerg Infect Dis 18:1983–1993. 12 doi:10.3201/eid1812.120875
- Prasad AN, Woolsey C, Geisbert JB, et al. 2020. Resistance of cynomolgus monkeys to nipah and hendra virus disease is associated with cell-mediated and humoral immunity. J Infect Dis 221:S436–S447. Supplement_4 doi:10.1093/infdis/jiz613
- Albariño CG, Wiggleton Guerrero L, Chakrabarti AK, et al. Transcriptional analysis of viral mRnas reveals common transcription patterns in cells infected by five different filoviruses. PLoS ONE. 2018 Aug 2;13(8):e0201827. doi:10.1371/journal.pone.0201827
- Guillaume V, Lefeuvre A, Faure C, et al. 2004. Specific detection of Nipah virus using real-time RT-PCR (TaqMan). J Virol Methods 120:229–237. 2 doi:10.1016/j.jviromet.2004.05.018
- Zivcec M, Safronetz D, Haddock E, et al. 2011. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus). J Immunol Methods 368:24–35. 1–2 doi:10.1016/j.jim.2011.02.004
- Culling CFA. 1974. Handbook of histopathological and histochemical techniques. 3rd ed. Butterworths. doi:10.1016/C2013-0-04011-X
- Ong S Tin, Yusoff K, Kho C Ling, Abdullah J Ong and Tan W Siang. (2009). Mutagenesis of the nucleocapsid protein of Nipah virus involved in capsid assembly. Journal of General Virology, 90(2), 392–397. doi:10.1099/vir.0.005710-0
- Chan YP, Koh CL, Lam SK, et al. 2004. Mapping of domains responsible for nucleocapsid protein–phosphoprotein interaction of henipaviruses. J Gen Virol 85:1675–1684. 6 doi:10.1099/vir.0.19752-0
- Omi-Furutani M, Yoneda M, Fujita K, et al. 2010. Novel phosphoprotein-interacting region in nipah virus nucleocapsid protein and its involvement in viral replication. J Virol 84:9793–9799. 19 doi:10.1128/JVI.00339-10
- Ranadheera C, Proulx R, Chaiyakul M, et al. 2018. The interaction between the Nipah virus nucleocapsid protein and phosphoprotein regulates virus replication. 1. Sci Rep 8:15994. 1 doi:10.1038/s41598-018-34484-7
- Shaw ML, García-Sastre A, Palese P, et al. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J Virol 78:5633–5641. 11 doi:10.1128/JVI.78.11.5633-5641.2004
- Ciancanelli MJ, Volchkova VA, Shaw ML, et al. 2009. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. J Virol 83:7828–7841. 16 doi:10.1128/JVI.02610-08
- Hagmaier K, Stock N, Goodbourn S, et al. 2006. A single amino acid substitution in the V protein of Nipah virus alters its ability to block interferon signalling in cells from different species. J Gen Virol 87:3649–3653. 12 doi:10.1099/vir.0.82261-0
- Yabukarski F, Lawrence P, Tarbouriech N, et al. 2014. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. 9. Nat Struct Mol Biol 21:754–759. 9 doi:10.1038/nsmb.2868
- Rodriguez JJ, Cruz CD, Horvath CM 2004. Identification of the nuclear export signal and STAT-Binding domains of the nipah virus v protein reveals mechanisms underlying interferon evasion. J Virol 78:5358–5367. 10 doi:10.1128/JVI.78.10.5358-5367.2004
- Bruhn JF, Hotard AL, Spiropoulou CF, et al. 2019. A conserved basic patch and central kink in the nipah virus phosphoprotein multimerization domain are essential for polymerase function. Structure 27:660–668.e4. 4 doi:10.1016/j.str.2019.01.012
- Patch JR, Han Z, McCarthy SE, et al. 2008. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J 5:137. 1 doi:10.1186/1743-422X-5-137
- Ciancanelli MJ, Basler CF 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 80:12070–12078. 24 doi:10.1128/JVI.01743-06
- Guillaume V, Aslan H, Ainouze M, et al. 2006. Evidence of a potential receptor-binding site on the Nipah Virus G Protein (NiV-G): identification of Globular Head Residues with a Role in Fusion Promotion and Their Localization on an NiV-G structural model. J Virol 80:7546–7554. 15 doi:10.1128/JVI.00190-06
- Negrete OA, Chu D, Aguilar HC, et al. 2007. Single amino acid changes in the Nipah and hendra virus attachment glycoproteins distinguish EphrinB2 from EphrinB3 usage. J Virol 81:10804–10814. 19 doi:10.1128/JVI.00999-07
- Weis M, Maisner A 2015. Nipah virus fusion protein: importance of the cytoplasmic tail for endosomal trafficking and bioactivity. Eur J Cell Biol 94:316–322. 7–9 doi:10.1016/j.ejcb.2015.05.005
- Tit-Oon P, Tharakaraman K, Artpradit C, et al. 2020. Prediction of the binding interface between monoclonal antibody m102.4 and Nipah attachment glycoprotein using structure-guided alanine scanning and computational docking. 1. Sci Rep 10:18256. 1 doi:10.1038/s41598-020-75056-y
- Chang L-Y, Mohd Ali A, Hassan SS, et al. 2006. Quantitative estimation of Nipah virus replication kinetics in vitro. Virol J 3:47. 1 doi:10.1186/1743-422X-3-47
- Whelan SPJ, Barr JN, Wertz GW 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119.
- Welch SR, Scholte FEM, Harmon JR, et al. 2020. In Situ imaging of fluorescent nipah virus respiratory and neurological tissue tropism in the syrian hamster model. J Infect Dis 221:S448–S453. Supplement_4 doi:10.1093/infdis/jiz393
- Rockx B, Brining D, Kramer J, et al. 2011. Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J Virol 85:7658–7671. 15 doi:10.1128/JVI.00473-11
- Wong KT, Grosjean I, Brisson C, et al. 2003. A golden hamster model for human acute Nipah virus infection. Am J Pathol 163:2127–2137. 5 doi:10.1016/S0002-9440(10)63569-9
- Mathieu C, Pohl C, Szecsi J, et al. 2011. Nipah virus uses leukocytes for efficient dissemination within a host ▿. J Virol 85:7863–7871. 15 doi:10.1128/JVI.00549-11
- Weingartl H, Czub S, Copps J, et al. 2005. Invasion of the central nervous system in a porcine host by Nipah virus. J Virol 79:7528–7534. 12 doi:10.1128/JVI.79.12.7528-7534.2005
- Munster VJ, Prescott JB, Bushmaker T, et al. 2012. Rapid Nipah virus entry into the central nervous system of hamsters via the olfactory route. Sci Rep 2:736. 1 doi:10.1038/srep00736
- Negrete OA, Levroney EL, Aguilar HC, et al. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. 7049. Nature 436:401–405. 7049 doi:10.1038/nature03838
- Negrete OA, Wolf MC, Aguilar HC, et al. 2006. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLOS Pathog 2:e7. 2 doi:10.1371/journal.ppat.0020007
- Baseler L, Scott DP, Saturday G, et al. 2016. Identifying early target cells of Nipah virus infection in Syrian Hamsters. PLoS Negl Trop Dis 10:e0005120. 11 doi:10.1371/journal.pntd.0005120
- Akira S, Hirano T, Taga T, et al. 1990. Biology of multifunctional cytokines: iL 6 and related molecules (IL 1 and TNF). Faseb J 4:2860–2867. 11 doi:10.1096/fasebj.4.11.2199284
- Junttila IS. Tuning the cytokine responses: an Update on interleukin (IL)-4 and IL-13 receptor complexes. Front Immunol. 2018;9. doi:10.3389/fimmu.2018.00888