ABSTRACT
Influenza A viruses (IAVs) pose a serious risk to both human and animal health. IAVs’ receptor binding characteristics account for a major portion of their host range and tissue tropism. While the function of neuraminidase (NA) in promoting the release of progeny virus is well-known, its role in the virus entry process remains poorly understood. Studies have suggested that certain subtypes of NA can act as receptor-binding proteins, either alone or in conjunction with haemagglutinin (HA). An important distinction is that NA from the avian influenza virus have a second sialic acid-binding site (2SBS) that is preserved in avian strains but missing in human or swine strains. Those observations suggest that the 2SBS may play a key role in the adaptation of the avian influenza virus to mammalian hosts. In this review, we provide an update of the recent research advances in the receptor-binding role of NA and highlight its underestimated importance during the early stages of the IAV life cycle. By doing so, we aim to provide new insights into the mechanisms underlying IAV host adaptation and pathogenesis.
Introduction
Effective transmission of influenza A viruses (IAVs) between hosts requires a delicate functional balance between haemagglutinin (HA) and neuraminidase (NA). HA is widely recognized that the receptor-binding protein HA initiates viral infection by interacting with sialic acid (SA) receptors on the surface of the host cell, whereas the receptor-cleaving protein NA facilitates the release of progeny virus from infected cells by cleaving SAs from cellular receptors [Citation1]. However, recent studies have provided increasing evidence that NA can also bind directly to receptors and play a crucial role in the early stages of viral infection. At first, it was found that H3N2 IAV, responsible for a pandemic in humans, could grow effectively in Vero cells, but not in chicken embryos. The progenitor virus produced in Vero cells was found to agglutinate human red blood cells (RBCs), but not chicken RBCs [Citation2]. Furthermore, post-infection ferret antiserum displayed poor haemagglutination inhibition activity against human H3N2 viruses isolated from MDCK cells [Citation3]. In a series of studies aiming to investigate these phenomena, it was observed that the HA receptor binding ability of H3N2 human influenza virus was reduced [Citation4], and its receptor-binding function was compensated by NA [Citation3]. It’s worth noting that some H3N2 IAV cultured in MDCK cells only rely on NA protein to agglutinate RBCs [Citation5]. Meanwhile, studies have highlighted that the N1, N2, and N9 subtypes of NA from avian influenza viruses contain a second sialic acid-binding site (2SBS). This 2SBS is located near the enzyme cleavage function site and can bind to SA receptors [Citation6], thus affecting the functional balance between HA and NA [Citation7]. These results have revealed fresh understandings of the function of NA and shed light on the functional equilibrium between HA and NA [Citation8].
IAVs and SA receptors
IAVs are enveloped viruses with an 8-segment negative-sense, single-stranded RNA genome that are members of the Orthomyxoviridae family [Citation9]. Seasonal flu caused by these viruses is responsible for a significant number of respiratory deaths globally each year, with estimated fatalities ranging from 290,000 to 650,000 [Citation10]. IAVs can generate both pandemic and seasonal epidemics and can infect a variety of hosts. Moreover, a devastating pandemic can result from the cross-species transmission of animal IAVs to humans. A “host jump” of the H1N1 avian influenza virus, for instance, is thought to have been the cause of the 1918 pandemic, the worst in recent history, which is estimated to have killed 40 million people worldwide [Citation11,Citation12].
Structurally, IAVs are typically spherical, filamentous, or elliptical in shape, and their nucleocapsids, housing their RNA genomes, are coated with a lipid bilayer membrane [Citation13]. The virus membrane is coated with two types of glycoproteins, HA and NA, which have distinct morphologies and functions. IAVs can be classified into different subtypes based on the antigenicity of their HA and NA proteins. Presently, researchers have identified 18 HA subtypes and 11 NA subtypes. In contrast to the H17N10 and H18N11 subtypes of IAV, which were discovered in bats [Citation14], the H1-H16 subtypes of IAV are commonly found in ducks [Citation15].
One of the main factors limiting the transmission of IAV between species is the receptor-binding property. Glycoproteins and glycolipids containing SA serve as receptors for IAV. The SA receptors are made up of the terminal SA residues, mannose, galactose, and N-acetylglucosamine [Citation16]. The glycosidic linkage between SA residues and downstream glycans plays a significant role in virus tropism. Generally, avian influenza viruses show a strong preference for SAα-2,3 Gal, while human influenza viruses prefer to bind SA attached to galactose through an α-2,6 linkage [Citation17]. IAVs need the receptor-binding site (RBS) on the head of their HA to recognize and attach to SA receptors, and changes to the RBS can change how specifically an IAV binds to a particular receptor. For example, the 2013 human H7N9 IAV acquired the Q226L and T160A mutations at the RBS, allowing it to bind to both SA-2,3 Gal and SA-2,6 Gal receptors [Citation18].
Influenza a virus and NA
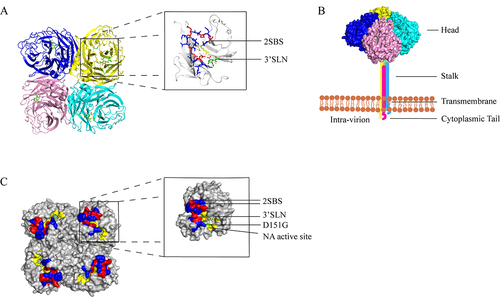
The NA is represented by mushroom-shaped spikes on the virion surface and is encoded by the sixth segment of the IAV genome [Citation19]. NA is a homo-tetramer protein with a molecular weight of 240 kDa, accounting for approximately 10–20% of the total glycoprotein on the virion surface (). Each polypeptide monomer has about 470 amino acid residues and consists of a globular head domain, a transmembrane region, and a highly conserved C-terminal cytoplasmic tail [Citation20,Citation21] (). Depletion or substitution of amino acid sequences (MNPNOK) of the NA cytoplasmic tails does not affect the enzyme activity; however, it is related to the assembly and budding of the virus [Citation22]. The transmembrane region of NA is an N-terminal hydrophobic transmembrane structure that connects NA to the virus envelope [Citation23]. The head domain of NA contains enzyme activity sites and antigenic epitopes [Citation24]. NA catalyzes the hydrolysis of glucoside bonds between the distal SA residues and the adjacent glycoproteins. The deletion of amino acid sites in the stalk of NA affects the enzyme activity and a shortened stalk may cause the head region to be unable to effectively act on the substrate [Citation25]. The absence of glycosylation sites in the stalk region of NA was found to enhance the virulence of IAV in mice [Citation26]. According to a recent study, NA activity fluctuates within a specific bandwidth rather than evolving to a single optimal value [Citation27]. The highly conserved NA enzyme active site makes an excellent target for the creation of brand-new anti-influenza medications.
Figure 1. (a) the structure of the NA protein tetrad displaying the head region is depicted. Each monomer is shown to have an SA residue attached to its head, which serves as the site of enzyme activity. (b) the four polypeptide monomers of NA fold and assemble into a tetrameric structure encompassing a spherical head region, a stem, a transmembrane region, and a cytoplasmic tail. The head region of the depicted NA was derived from the Protein Data Bank (PDB:4GZW), and the corresponding protein model was generated using Pymol software. (c) the relative positions of the enzyme active site (highlighted in yellow) and the second receptor binding site (highlighted in blue and red: 366–373, 399–404, 430–433) at the head region of each NA monomer are shown, with the red area (364, 367, 369, 400, 403, 432) denoting the contact site of the second receptor binding site of NA. The location of the D151G mutation is illustrated in green. Each monomer binds to an α-2,3 sialic acid (SA). The NA structure displayed in the figure was obtained from the Protein Data Bank (PDB:4GZW), and the protein model was generated using Pymol software.
Function of NA protein
It is widely accepted that NA, by acting as a counterbalance to HA, plays a critical role in the lifecycle of IAVs. Firstly, NA removes SA from respiratory mucin cilia and cellular glycoprotein envelopes, thereby facilitating the virus’s migration through respiratory mucus [Citation28]. Secondly, NA catalyzes the cleavage of terminal SAs from virus receptors, which promotes virus fusion with the host cell membrane [Citation29]. Thirdly, during the final viral budding process, NA removes SA receptors on the cell surface to aid in the release of progeny virus, which can then infect neighbouring cells [Citation30]. Although the corresponding HA of human IAVs has a strong preference for SAα-2,6 Gal, NA from both avian and human IAVs hydrolyse SAα-2,3 Gal more efficiently. However, the difference in NA enzyme activity between α-2,3 and α-2,6 linkages is relatively smaller in human IAVs than in avian IAVs [Citation31]. In addition to its ability to catalyse the hydrolysis of SA receptors, some subtypes of NA proteins (N1, N2, and N9) from avian IAVs have also been found to possess haemagglutination activity, which is related to the second sialic acid-binding site (2SBS) of the NA proteins [Citation32].
Recent research suggests that NA is engaged in the early phases of the IAVs lifecycle in addition to its enzymatic activities by encouraging virus entrance. Ohuchi et al. discovered that NA inhibitors greatly reduced IAV susceptibility to MDCK cells, showing that NA is important in increasing virus entry and enhancing infection efficiency [Citation33]. This was further substantiated by a recent study showing that NA protein can increase the effectiveness of viral infection by encouraging recurrent endocytosis, virus release on the cell surface, and virus motility [Citation34]. Similar to these observations, other research has demonstrated that NA encourages virus rolling on the cell surface, improving the effectiveness of virus attaching to host cells during the early stages of infection [Citation35]. However, the virus motility idea is still not widely recognized, and it is still unknown what molecular process underlies the HA-NA balance in IAV movement patterns.
NA and the second sialic acid-binding site (2SBS)
Laver et al. first discovered in 1984 that N9 from avian influenza viruses could haemagglutinate animal RBCs, whereas neither HA nor NA antibodies alone could inhibit viral haemagglutination to animal RBCs [Citation36]. Webster et al. proposed that the haemagglutination activity of N9 was associated with the presence of 2SBS near the enzyme activity site of NA () [Citation32]. They further demonstrated that mutations in the loops of 368–370 and 399–403 of N9 2SBS resulted in a loss of haemagglutination activity [Citation32]. Meanwhile, the N2 A/Tokyo/3/67 haemagglutinin was shown to be able to haemagglutinate when the amino acids in its loops of 368–370 and 399–403 were switched out for those of the N9 haemagglutinin [Citation37]. These studies clearly suggested that N9 had 2SBS, and that the loops of 368–370 and 399–403 in the NA contained key residues that interacted with the SA. However, the molecular mechanism of the 2SBS-SA interaction remains largely unknown. In 1995, Air et al. reported that the binding of 2SBS of NA to Neu5Ac was not sensitive to the enzyme activity of N9 NA, but could only be cut by bacterial sialidases [Citation38]. However, the structural evidence of 2SBS and the binding basis of 2SBS to SA were not revealed until 1997. Varghese et al. constructed the X-ray crystal structure of N9 from an avian influenza virus and demonstrated the existence of 2SBS capable of binding SA near the enzyme-active site in the head region of the N9 [Citation39]. The enzyme active site was found to connect with SA in a boat conformation, whereas the 2SBS interacted with SA through a chair conformation, according to prior structural investigations of NA-SA interactions [Citation39]. Most recently, Stefano Elli et al. investigated the atomic interactions between the 2SBS of avian-origin NAs and SA using molecular dynamics simulations. Their findings indicated that BM18 binds both human and avian ligands with comparable efficiency [Citation40].
Hemadsorption activity varies between NA subtypes
In 1995, Hausmann et al. demonstrated that N1 from A/FPV/Rostock/34 (H7N1) expressed in Sf9 cells exhibited haemagglutination activity towards chicken red blood cells [Citation41]. Interestingly, the haemagglutination activity of N1 was not affected by the NA enzyme activity inhibitor, indicating that N1 has distinct 2SBS compared to N2 [Citation41]. The haemadsorption capacity of NA, however, differs between subtypes. For instance, N1 binds to RBCs at both 4°C and 37°C, whereas N9 agglutinates chicken or human RBCs at 4°C but not at 37°C [Citation36,Citation41]. It is worth noting that the NAs from avian isolates had much higher hemadsorption activity than those from swine and human isolates [Citation42].
Function of 2SBS in IAV lifecycle
Due to the underestimating of NA’s significance in the early stages of the IAV life cycle, the function of 2SBS of NA has been poorly known for a very long time. The binding of 2SBS of NA to fetuin, however, contributes to the activity of the NA enzyme, whereas the binding of monovalent substrates has no influence on the NA enzyme activity, according to a recent study by Du et al. [Citation43]. Interestingly, mutations at non-SA contact residues 368 or 369 in loop 370 had an even greater impact than mutations at residue 372, which directly contacts SA [Citation43]. Moreover, Elli et al revealed that the substitution of S369 influences the binding of 6’SLN-LC but not 3’SLN-LC, indicating that the substitution at this location may dictate the binding specificity of 2SBS for the linkage type of Neu5Ac-Gal [Citation40]. Recent research has demonstrated that the H1N1 pandemic strain’s 2SBS mutations improve the ability of NA proteins to bind to polyvalent substrates, with variable impacts on NA binding to 2,3 or 2,6 SA [Citation43]. In addition, a Brownian dynamics (BD) simulation study discovered that the 2SBS’s electrostatic interaction with the avian influenza virus’s active site of NA plays a critical role in both the recognition of NA and SA receptors as well as the enzyme digesting function of NA [Citation44]. The HA-NA receptor balance necessary for effective viral replication depends on both the receptor-binding and receptor-destroying abilities of NA and its 2SBS [Citation45].
2SBS promote the interspecies transmission of H7N9 avian influenza virus
Dai et al. discovered that the receptor-binding property of NA from human H7N9 remained unaltered when using monovalent substrates, compared to the prior avian H7N9 that only infected birds. However, when the multivalent fetoprotein substrates were used for analysis, the binding of human H7N9 NA to the substrates was decreased due to a T401A mutation in the non-contact binding site of 2SBS [Citation46]. The Spanish strain N9 T401A has been observed to reduce both its NA and multivalent substrate-binding ability and NA enzyme activity, while the Anhui strain N9 A401T increased these same characteristics [Citation46]. It is essential to consider the consistent changes in the NA enzyme activity and binding ability to polyvalent substrates.
Benton et al. compared the binding of the two SA receptors to the H7N9 subtype IAV with or without 2SBS in the presence or absence of NA enzyme inhibitors [Citation6]. It was found that, with or without 2SBS, NA had a much weaker ability to bind to α-2,3 SA receptors in the absence of NA enzyme inhibitors than in the presence of them. This may be attributed to the strong cleavage capacity of N9 NA to α-2,3 SA receptors. For α-2,6 SA receptor, however, NA with 2SBS showed a stronger binding ability to α-2,6 SA receptor in a very short time when the NA enzyme inhibitor was not present. Their results demonstrated that NA binding to α-2,6 SA in H7N9 IAV exhibited a short-term enhancement only when both the NA alpha and the enzyme active site were present [Citation6]. This short-term enhancement helps the virus remain in the upper respiratory tract, which is abundant in α-2,6 SA receptors, for longer, thus increasing the possibility of it infecting humans. However, the NA enzyme activity centre of H7N9 IAV has a strong capacity to cleave avian α-2,3 SA receptors and a weak capacity to cleave human α-2,6 SA receptors [Citation6]. The release of progeny virions from the respiratory tract of humans is blocked, which prevents the spread of avian influenza viruses. This is related to the balance between HA and NA receptors. Additionally, Benton et al. found that the second-binding site (2SBS) of H7N9 NA receptor in the enzyme activity centre can promote the cleavage of α-2,6 SA receptor to some extent, thereby inhibiting the infection of the virus. Consequently, virus proliferation in the upper respiratory tract of humans is blocked after the release of the newborn virus particles, thus preventing the spread of avian influenza virus in humans. This is still related to the balance between HA and NA receptors. Moreover, Benton et al. also demonstrated that the 2SBS of H7N9 NA receptor could promote the cleavage of α-2,6 SA receptor in the enzyme activity centre to a certain degree [Citation6]. Once the HA and NA of avian influenza H7N9 reach a balance in their binding and cleavage of human α-2,6 SA receptors, it may help the virus adapt to humans. Recently, it was found that 2SBS and other potential low-affinity binding sites could increase the variety of heterogeneous multivalent interaction networks, which might then impact the effectiveness of NA binding and cleavage, affecting the virus’s mobility and release [Citation47]. Liu et al. demonstrated that hetero-multivalent binding significantly increased the efficiency of virus entry. These findings contradict the long-held notion that the avian α-2,3 SA receptor has little to no effect on human IAV infection, which requires α-2,6 SA binding [Citation47].
2SBS and interspecies transmission of IAV
Mikhail N and his team found that the 2SBS locus is preserved in viruses that have a high affinity for the α-2,3 receptor. However, avian viruses that mainly bind to the α-2,6 receptor (such as human, swine, and avian H9N2 viruses) have mutations in critical positions of this locus [Citation48]. Further investigations have shown that the conservation of 2SBS is only absent in H9N2 subtype viruses for N2 avian viruses [Citation7]. These results suggest that the functional impairment of 2SBS may be a crucial adaptation to maintain the HA-NA equilibrium for a pandemic virus.
In contrast to the human H3N2 virus, the sequences of the N2 virus’s 2SBS from birds and mammals were found to be conserved by Varghese et al. [Citation39]. Kobasa et al. detected most of the NA subtypes in poultry, swine, horses, and human IAV, and they observed that the haemagglutination activity of human N1 and N2 NA was significantly lower than that of avian N1 and N2 NA [Citation42]. Furthermore, the residues of N1 and N2 that interact with SA receptors at the 2SBS were highly conserved in avian influenza viruses but less conserved in human influenza viruses () [Citation42]. Uhlendorff et al. demonstrated that the haemagglutinin activity of the H2N2 human pandemic strain could be restored by introducing a point mutation (N367S) to the 2SBS [Citation49]. These results imply that avian IAV adaptation to humans may not be supported by the preserved 2SBS of N2 NA. Consequently, further research is required to fully understand the mechanism by which the NA 2SBS contributes to the avian influenza virus’ ability to cross the host barrier.
Figure 2. The sequence of the three loops (residues 366 to 373), 400 loop (residues 399 to 404), and 430 loop (residues 430 to 433) that form the second receptor binding site of the N1, N2, and N9 in humans and birds. This Seqlogo was generated by the WebLogo program (http://weblogo.berkeley.edu/logo.cgi) using strains retrieved from the GenBank database.

NA as a receptor binding protein
Hooper et al. artificially created HA-receptor-binding defective mutants in the laboratory, and their results showed that the G147R NA protein could compensate for the deficiency of HA receptor in the alternative HA-combining-ability-deficient type of mutant viruses as the receptor protein, yet still requiring HA-mediated virus-cell membrane fusion [Citation50]. Furthermore, genetic sequencing of human (H1N1) and avian (H5N1) IAVs has revealed that similar mutations occur naturally [Citation50]. Interestingly, G147R is situated above the NA enzyme active site, rather than directly at the 2SBS or NA enzyme active site [Citation50]. Consequently, Hooper et al. speculated that this mutation would not affect the enzyme activity of NA. They coupled human H1N1 IAV and avian H5N1 IAV NA mutant G147R in the lab and discovered that the pathogenicity in mice remained unaffected, as well as the NA protein activity of the monovalent substrate MUNANA [Citation51]. Ting-Hsuan Chen et al. recently discovered that the 2SBS 370 ring mutation-induced increase or reduction in NA enzyme activity may be related to the rise or decrease in HA titre [Citation52]. This implies that the influenza virus’s NA enzyme activity impacts the HA-NA balance, and changes to the ratio of HA to NA on the surface of the virus particle or the ratio of HA to NA receptors may impair the virus’s capacity to adapt to a new host.
The role of NA in receptor binding of recent human H3N2 virus
The subtype H3N2 of IAVs is one of the major IAV subtypes (H1N1, H3N2) that cause seasonal influenza epidemics in humans. Mutations near the Receptor Binding Site (RBS) on the HA head have been observed to drive the evolution of recent H3N2 IAVs, leading to a lower protection efficiency of the seasonal influenza vaccines than that of the H1N1 subtype [Citation53]. Furthermore, it is not uncommon for recent human H3N2 isolates to have decreased binding affinity to both. This is brought on by adaptive changes in the HA RBS, which also cause human H3N2 isolates to develop slowly in chicken embryos [Citation54]. There is mounting evidence that human-adapted N2 can supplement or even take the place of HA in the function of receptor-binding proteins.
In 1998, Grassauer et al. discovered that human IAV A/Vienna/47/96 and A/Vienna/81/96 of the H3N2 subtype, which were isolated from the Vienna pandemic, grew efficiently in Vero cells, but not in chicken embryos, and the progenies from Vero cells were unable to agglutinate chicken erythrocytes [Citation2]. This loss of haemagglutination activity indicated a decreased virus binding to SA receptors for H3N2 human IAV. Since then, a series of studies have been conducted to investigate the molecular mechanisms behind the changes in the binding properties of H3N2 IAVs. Lu et al. found that the HA gene G186V or A196T mutation of human H3N2 A/Singapore/21/04, which could not proliferate in chicken embryos, could enhance the replication ability of the virus in chicken embryos [Citation55]. Nobusawa et al. generated an A/Aichi/51/92 (H3N2) HA E190D mutant virus through site-directed mutagenesis and determined that the E190D mutation was the cause of the loss of H3’s binding ability to CRBC [Citation56]. By passing H3N2 virus without its haemagglutinin activity into MDCK cells and screening out the phenotypic reversing strain, it was found that V226I of H3 increased the affinity between HA and α-2,3 SA receptors, while S193R and L194I mutations enhanced the affinity between HA and α-2,6 SA receptor [Citation4]. These findings suggest that these H3 mutations lead to decreased binding of IAV to α-2,3 and α-2,6 SA receptors, indicating that NA may have been gradually assuming the role of receptor-binding proteins at this time.
Gulati et al. conducted a study to analyse the receptor-binding characteristics of H3N2 human influenza viruses from 1968 to 2012 and found that each year the viruses exhibited different receptor-binding characteristics [Citation57]. For example, the 2006, 2010, and 2012 pandemic influenza viruses employed NA to bind to α-2,3 SA receptors [Citation57]. These changes in receptor-binding properties did not adversely affect the pathogenicity or transmissibility of the virus in mammals. Mohr et al. studied H3N2 human influenza viruses isolated since 1994 and cultured in canine embryonic kidney cells (MDCK) and found that they relied solely on NA proteins to agglutinate RBCs [Citation5]. Mogling et al. investigated the changes in haemagglutination activity of H3N2 human influenza virus from 2000 to 2016 and discovered that the virus gradually lost its ability to agglutinate Turkey RBCs in the presence of oseltamivir inhibitors after 2003, and by 2012, almost all H3N2 viruses examined had lost their haemagglutination activity in the presence of Oseltamivir [Citation58]. This indicates that since 2003, the haemagglutination function of human H3N2 IAV has been increasingly mediated by NA rather than HA.
Mogling et al. discovered that the NA H150R located near the neuraminidase catalytic site was responsible for the increased haemagglutinating activity of NA, making haemagglutination inhibition assays useless for the selection of vaccine strains [Citation58]. Lin et al. found that the haemagglutination activity of H3N2 human influenza virus isolated from 2005 to 2009 could not be inhibited by HA antibody. This was because mutations, such as NA D151G, caused NA to become a receptor binding protein, which could not be cleaved by NA itself, but only lysed by certain bacterial sialases [Citation3]. The haemagglutination activity of N2 can be inhibited by NA inhibitors, such as Oseltamivir, but not neutralized by anti-HA antibodies [Citation3]. Zhu et al. compared the catalytic activity of the wild-type H3N2 human influenza virus NA with its D151G mutant using sugar microchips and other techniques, and found that the enzyme activity of the D151G mutant was significantly decreased, and the affinity of the D151G mutant with α-2,3 and α-2,6 SA receptors was stronger than that of the wild-type H3N2 IAV [Citation59]. They introduced D151G mutations into three H1N1 IAV strains (two human influenza strains and one swine influenza virus) and one human H3N2 IAV, and observed significant reductions in NA enzyme activity in all four strains [Citation59]. Interestingly, H150R and D151G mutations are located in the 150 loop of NA, at the edge of the active site of the enzyme, rather than at the 2SBS of NA [Citation3], suggesting that mutations in the 2SBS are not the only ones that can alter the NA receptor binding characteristics.
However, a study by Xue and others has demonstrated that direct sequencing of H3N2 influenza viruses from clinical samples has not revealed the D151G mutation, which is mainly due to the low-level mutation caused by the passage of clinical samples in MDCK cells [Citation60]. Brown and colleagues conducted an analysis of 151 North American variations of clinical A/H3N2 samples using NGS. Their findings confirmed the notion that these variations form independently during cell culture proliferation [Citation61]. The study also discovered that passage in HAE cells might abolish the NA-mediated binding of A/H3N2 isolates, which was seen as an artefact of MDCK/MDCk-SIAT1 cell passage [Citation61]. The distribution of MDCK cells may be distinct from that of the human respiratory tract receptors, but the precise reasons remain to be explored. These findings also imply that the separation and culture of MDCK cells or chicken embryos may lead to genetic information bias, highlighting the significance of direct sequencing from clinical specimens.
Mutations in functional regions of NA receptors and cross-species transmission
Cross-host infection with the avian influenza subtypes H7N9, H9N2, H10N8, H5N6, and others occasionally happens and constitutes a severe threat to public health [Citation62–64]; fortunately, however, these viruses do not yet possess an effective human-to-human transmission capability. If they were to succeed in breaching the host barrier, they would undoubtedly cause even greater harm to public health. Adaptation of IAV to new hosts necessitates the adjustment of NA and HA functional balance in order to replicate effectively in the new host. DE Vries et al. proposed that the potential impact of NA on the motion of IAV particles associated with receptors has been previously undervalued [Citation65]. NA may prevent the virus from being caught by the bait receptor when it passes through the mucosal layer of the respiratory system, according to some theories. Contrarily, it is believed that NA maintains a functional equilibrium with HA and is essential for the virus’s ability to connect to the host receptor. Mutations in NA receptor functional regions can lead to NA replacing HA as a receptor binding protein and may affect the virus’s affinity to different receptors. Recently, Du et al. discovered that the H5N1 avian influenza virus NA 2SBS, K432E mutation may interfere with the function of the HA and NA balance and influence viral replication in order to adapt to a new host and replicate efficiently [Citation66]. In a recent study by Meiling Dai and colleagues [Citation27], it was observed that replacement of K432E in 2SBS (A/North Carolina/07/2013, NC/13 NA) resulted in very low NA activity, particularly for multivalent substrates. According to this, NA activity against human viruses may be improved by low-affinity binding at this location. Moreover, it has been demonstrated that the compensatory mutation S223N in the HA gene increases the affinity for human receptors, facilitating the transmission of avian IAV among other species [Citation66]. The 2SBS mutation, which increased binding to the human −2,6 SA receptor and decreased binding to the avian −2,3 SA receptor, was found to have occurred in H9N2 avian IAV before the HA mutation, according to phylogenetic analysis [Citation66]. These results suggest that the H9N2 avian influenza virus’s host tropism may be significantly altered by NA mutations. Se-Hee An et al. discovered that the mutation of NA 2SBS preceded the HA Q226L mutation, implying that H9N2 AIV may initially reduce the affinity of the avian receptor to NA, leading to decreased enzymatic activity and thereby driving the HA mutation to boost the affinity to the human receptor [Citation67]. Through a genetic evolutionary analysis, Dai et al. discovered that the NA T401A mutation of H7N9 occurred prior to the adaptive mutations of the HA gene Q226L and T160A in mammals, leading them to conclude that the mutation of 2SBS may have promoted a change in the HA receptor binding characteristics, thereby facilitating the cross-species transmission of the H7N9 virus [Citation46].
Using glycan microarray experiments, it was found that NA binding to human-type-like receptors, primarily due to the S430G mutation located near the 2SBS, may be independent of the sialidase pocket [Citation68]. Additionally, NA mutations in human-like H5N1 strains (A46D, L204M, S319F, and S430G) were found to reduce the activity and expression level of NA, while enhancing the binding affinity of HA to the human-type receptor through a synergistic effect. These findings imply that the avian influenza virus’s mutation of the HA receptor binding site enhances the virus’s binding affinity to the human −2,6 SA receptor while decreasing the virus’s binding affinity to the avian −2,3 SA receptor, increasing the virus’s transmission to humans. Importantly, the mutation in the NA receptor functional region may drive the HA mutation, highlighting the essential role of NA receptor functional region mutations in promoting the “host jump” of avian influenza virus. It is important to note that these results are distinct from any previously published research by the author. A summary of the molecular markers associated with receptor-binding properties of NA was shown in .
Table 1. Molecular markers associated with receptor-binding properties of NA.
Concluding remarks
Concerns regarding the potential of a pandemic have been raised by cases of human infection with avian influenza viruses, including the H5N1, H5N6/N8, H7N9, H3N8, H10N8, and H9N2 subtypes. The effect of the HA protein on the capacity of viruses to switch host species has been thoroughly investigated by scientists. Recent studies have shown that mutations in the 2SBS region may alter the receptor-binding properties of the NA protein, affecting the avian influenza virus’s ability to adapt to the host. Similarly, mutations in the NA protein of human influenza virus subtypes H1N1, H2N2, and H3N2 also affect receptor-binding characteristics. These findings provide a new perspective on preventing and treating influenza. Changes in the HA-NA balance with respect to host receptors can result from mutations in the 2SBS region, which can then alter the receptor-binding properties of related HA proteins. The biological importance of the NA 2SBS region, the molecular mechanism of NA binding to SA receptors, and the mechanism of NA 2SBS regulating the complex and dynamic balance between HA, NA, and SA receptors remain largely unknowns despite the fact that these mutations have been occurring frequently and going unnoticed for a while. Further exploration by scholars is necessary to understand the cross-host transmission, pathogenesis, and prevention of the NA receptor binding property of influenza virus.
Acknowledgements
We thank Dr. Jinyue Guo, Foshan University, for the valuable discussions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Ward MJ, Lycett SJ, Avila D, et al. Evolutionary interactions between haemagglutinin and neuraminidase in avian influenza. BMC Evol Biol. 2013;13(1):222. doi: 10.1186/1471-2148-13-222
- Grassauer A, Egorov AY, Ferko B, et al. A host restriction-based selection system for influenza haemagglutinin transfectant viruses. J Gen Virol. 1998;79(Pt 6):1405–11. doi: 10.1099/0022-1317-79-6-1405
- Lin YP, Gregory V, Collins P, et al. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol. 2010;84(13):6769–6781. doi: 10.1128/JVI.00458-10
- Medeiros R, Escriou N, Naffakh N, et al. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology. 2001;289(1):74–85. doi: 10.1006/viro.2001.1121
- Mohr PG, Deng YM, McKimm-Breschkin JL. The neuraminidases of MDCK grown human influenza A(H3N2) viruses isolated since 1994 can demonstrate receptor binding. Virol J. 2015;12(1):67. doi: 10.1186/s12985-015-0295-3
- Benton DJ, Wharton SA, Martin SR, et al. Role of neuraminidase in influenza A(H7N9) virus receptor binding. J Virol. 2017;91(11): Virol 91. doi: 10.1128/JVI.02293-16
- Du W, Guo H, Nijman VS, et al. The 2nd sialic acid-binding site of influenza a virus neuraminidase is an important determinant of the hemagglutinin-neuraminidase-receptor balance. PLOS Pathog. 2019;15(6):e1007860. doi: 10.1371/journal.ppat.1007860
- Wen F, Wan XF. Influenza neuraminidase: Underrated role in receptor binding. Trends Microbiol. 2019;27(6):477–479. doi: 10.1016/j.tim.2019.03.001
- Peiris JS, Poon LL, Guan Y. Emergence of a novel swine-origin influenza a virus (S-OIV) H1N1 virus in humans. J Clin Virol. 2009;45(3):169–173. doi: 10.1016/j.jcv.2009.06.006
- Iuliano AD, Roguski KM, Chang HH, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. doi: 10.1016/S0140-6736(17)33293-2
- Johnson NP, Mueller J. Updating the accounts: Global Mortality of the 1918-1920 “Spanish” influenza pandemic. Bulletin Of The History Of Medicine. 2002;76(1):105–115. doi: 10.1353/bhm.2002.0022
- Taubenberger JK. The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc. 2006;150:86–112. 1
- Nayak DP, Balogun RA, Yamada H, et al. Influenza virus morphogenesis and budding. Virus Res. 2009;143(2):147–161. doi: 10.1016/j.virusres.2009.05.010
- Wu Y, Wu Y, Tefsen B, et al. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014;22(4):183–191. doi: 10.1016/j.tim.2014.01.010
- Shi Y, Wu Y, Zhang W, et al. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza a viruses. Nat Rev Microbiol. 2014;12(12):822–831. doi: 10.1038/nrmicro3362
- Long JS, Mistry B, Haslam SM, et al. Host and viral determinants of influenza a virus species specificity. Nat Rev Microbiol. 2019;17(2):67–81. doi: 10.1038/s41579-018-0115-z
- Ge S, Wang Z. An overview of influenza a virus receptors. Crit Rev Microbiol. 2011;37(2):157–165. doi: 10.3109/1040841X.2010.536523
- Zhou J, Wang D, Gao R, et al. Biological features of novel avian influenza a (H7N9) virus. Nature. 2013;499(7459):500–503. doi: 10.1038/nature12379
- Bossart-Whitaker P, Carson M, Babu YS, et al. Three-dimensional structure of influenza a N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-Acetyl neuraminic acid. J Mol Biol. 1993;232(4):1069–1083. doi: 10.1006/jmbi.1993.1461
- McAuley JL, Gilbertson BP, Trifkovic S, et al. Influenza virus neuraminidase structure and functions. Front Microbiol. 2019;10:39. doi: 10.3389/fmicb.2019.00039
- Blok J, Air GM. Variation in the membrane-insertion and “stalk” sequences in eight subtypes of influenza type a virus neuraminidase. Biochemistry. 1982;21(17):4001–4007. doi: 10.1021/bi00260a015
- Barman S, Adhikary L, Chakrabarti AK, et al. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J Virol. 2004;78(10):5258–5269. doi: 10.1128/JVI.78.10.5258-5269.2004
- Ernst AM, Zacherl S, Herrmann A, et al. Differential transport of Influenza a neuraminidase signal anchor peptides to the plasma membrane. FEBS Lett. 2013;587(9):1411–1417. doi: 10.1016/j.febslet.2013.03.019
- Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303(5912):41–44. doi: 10.1038/303041a0
- Blumenkrantz D, Roberts KL, Shelton H, et al. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J Virol. 2013;87(19):10539–10551. doi: 10.1128/JVI.00967-13
- Park S, Il Kim J, Lee I, et al. Adaptive mutations of neuraminidase stalk truncation and deglycosylation confer enhanced pathogenicity of influenza a viruses. Sci Rep. 2017;7(1):10928. doi: 10.1038/s41598-017-11348-0
- Dai M, Du W, Martínez-Romero C, et al. Analysis of the evolution of pandemic influenza A(H1N1) virus neuraminidase reveals entanglement of different phenotypic characteristics. MBio. 2021;12: doi: 10.1128/mBio.00287-21
- Matrosovich MN, Matrosovich TY, Gray T, et al. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78(22):12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004
- Yang X, Steukers L, Forier K, et al. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS One. 2014;9(10):e110026. doi: 10.1371/journal.pone.0110026
- Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12(3):159–166. doi: 10.1002/rmv.352
- Li Y, Cao H, Dao N, et al. High-throughput neuraminidase substrate specificity study of human and avian influenza a viruses. Virology. 2011;415(1):12–19. doi: 10.1016/j.virol.2011.03.024
- Webster RG, Air GM, Metzger DW, et al. Antigenic structure and variation in an influenza virus N9 neuraminidase. J Virol. 1987;61(9):2910–2916. doi: 10.1128/jvi.61.9.2910-2916.1987
- Ohuchi M, Asaoka N, Sakai T, et al. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 2006;8(5):1287–1293. doi: 10.1016/j.micinf.2005.12.008
- Sakai T, Nishimura SI, Naito T, et al. Influenza a virus hemagglutinin and neuraminidase act as novel motile machinery. Sci Rep. 2017;7(1):45043. doi: 10.1038/srep45043
- Guo H, Rabouw H, Slomp A, et al. Kinetic analysis of the influenza a virus HA/NA balance reveals contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces. PLOS Pathog. 2018;14(8):e1007233. doi: 10.1371/journal.ppat.1007233
- Laver WG, Colman PM, Webster RG, et al. Influenza virus neuraminidase with hemagglutinin activity. Virology. 1984;137(2):314–323. doi: 10.1016/0042-6822(84)90223-X
- Nuss JM, Air GM. Transfer of the hemagglutinin activity of influenza virus neuraminidase subtype N9 into an N2 neuraminidase background. Virology. 1991;183(2):496–504. doi: 10.1016/0042-6822(91)90979-L
- Air GM, Laver WG. Red cells bound to influenza virus N9 neuraminidase are not released by the N9 neuraminidase activity. Virology. 1995;211(1):278–284. doi: 10.1006/viro.1995.1401
- Varghese JN, Colman PM, van Donkelaar A, et al. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci U S A. 1997;94(22):11808–11812. doi: 10.1073/pnas.94.22.11808
- Elli S, Gambacorta N, Rudd TR, et al. MD simulation of the interaction between sialoglycans and the second sialic acid binding site of influenza a virus N1 neuraminidase. Biochem J. 2021;478(2):423–441. doi: 10.1042/BCJ20200670
- Hausmann J, Kretzschmar E, Garten W, et al. N1 neuraminidase of influenza virus A/FPV/Rostock/34 has haemadsorbing activity. J Gen Virol. 1995;76(Pt 7):1719–1728. doi: 10.1099/0022-1317-76-7-1719
- Kobasa D, Rodgers ME, Wells K, et al. Neuraminidase hemadsorption activity, conserved in avian influenza a viruses, does not influence viral replication in ducks. J Virol. 1997;71(9):6706–6713. doi: 10.1128/jvi.71.9.6706-6713.1997
- Du W, Dai M, Li Z, et al. Substrate binding by the second sialic acid-binding site of influenza a virus N1 neuraminidase contributes to enzymatic activity. J Virol. 2018;92(20). doi: 10.1128/JVI.01243-18
- Sung JC, Van Wynsberghe AW, Amaro RE, et al. Role of secondary sialic acid binding sites in influenza N1 neuraminidase. J Am Chem Soc. 2010;132(9):2883–2885. doi: 10.1021/ja9073672
- Du W, de Vries E, van Kuppeveld FJM, et al. Second sialic acid-binding site of influenza a virus neuraminidase: binding receptors for efficient release. FEBS J. 2021;288:5598–5612. doi: 10.1111/febs.15668
- Dai M, McBride R, Dortmans J, et al. Mutation of the second sialic acid-binding site, resulting in reduced neuraminidase activity, preceded the emergence of H7N9 influenza a virus. J Virol 91. 2017;91(9). doi: 10.1128/JVI.00049-17
- Liu M, Huang LZX, Smits AA, et al. Human-type sialic acid receptors contribute to avian influenza a virus binding and entry by hetero-multivalent interactions. Nat Commun. 2022;13:4054. doi: 10.1038/s41467-022-31840-0
- Matrosovich MN, Krauss S, Webster RG. H9N2 influenza a viruses from poultry in asia have human virus-like receptor specificity. Virology. 2001;281(2):156–162. doi: 10.1006/viro.2000.0799
- Uhlendorff J, Matrosovich T, Klenk HD, et al. Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch Virol. 2009;154(6):945–957. doi: 10.1007/s00705-009-0393-x
- Hooper KA, Bloom JD. A mutant influenza virus that uses an N1 neuraminidase as the receptor-binding protein. J Virol. 2013;87(23):12531–12540. doi: 10.1128/JVI.01889-13
- Hooper KA, Crowe JE Jr., Bloom JD, et al. Influenza viruses with receptor-binding N1 neuraminidases occur sporadically in several lineages and show no attenuation in cell culture or mice. J Virol. 2015;89(7):3737–3745. doi: 10.1128/JVI.00012-15
- Chen TH, Chen CC, Wu SC. Neuraminidase (NA) 370-loop mutations of the 2009 pandemic H1N1 viruses affect NA enzyme activity, hemagglutination titer, mouse virulence, and inactivated-virus immunogenicity. Viruses. 2022;14(6):1304. Viruses 14. doi: 10.3390/v14061304
- Koel BF, Burke DF, Bestebroer TM, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342(6161):976–979. doi: 10.1126/science.1244730
- Parker L, Wharton SA, Martin SR, et al. Effects of egg-adaptation on receptor-binding and antigenic properties of recent influenza a (H3N2) vaccine viruses. J Gen Virol. 2016;97(6):1333–1344. doi: 10.1099/jgv.0.000457
- Lu B, Zhou H, Chan W, et al. Single amino acid substitutions in the hemagglutinin of influenza A/Singapore/21/04 (H3N2) increase virus growth in embryonated chicken eggs. Vaccine. 2006;24(44–46):6691–6693. doi: 10.1016/j.vaccine.2006.05.062
- Nobusawa E, Ishihara H, Morishita T, et al. Change in receptor-binding specificity of recent human influenza a viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278(2):587–596. doi: 10.1006/viro.2000.0679
- Gulati S, Smith DF, Cummings RD, et al. Human H3N2 influenza viruses isolated from 1968 to 2012 show varying preference for receptor substructures with no apparent consequences for disease or spread. PLoS One. 2013;8(6):e66325. doi: 10.1371/journal.pone.0066325
- Mögling R, Richard MJ, Vliet SV, et al. Neuraminidase-mediated haemagglutination of recent human influenza A(H3N2) viruses is determined by arginine 150 flanking the neuraminidase catalytic site. J Gen Virol. 2017;98(6):1274–1281. doi: 10.1099/jgv.0.000809
- Zhu X, McBride R, Nycholat CM, et al. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors. J Virol. 2012;86(24):13371–13383. doi: 10.1128/JVI.01426-12
- Xue KS, Greninger AL, Pérez-Osorio A, et al. Cooperating H3N2 influenza virus variants are not detectable in primary clinical samples. mSphere. 2018;3(1). doi: 10.1128/mSphereDirect.00552-17
- Brown JC, Barclay WS, Galiano M, et al. Passage of influenza A/H3N2 viruses in human airway cells removes artefactual variants associated with neuraminidase-mediated binding. J Gen Virol. 2020;101(5):456–466. doi: 10.1099/jgv.0.001348
- Almayahi ZK, Al Kindi H, Davies CT, et al. First report of human infection with avian influenza A(H9N2) virus in Oman: The need for a one health approach. Int J Infect Dis. 2020;91:169–173. doi: 10.1016/j.ijid.2019.11.020
- Bi Y, Mei K, Shi W, et al. Two novel reassortants of avian influenza a (H5N6) virus in China. J Gen Virol. 2015;96(5):975–981. doi: 10.1099/vir.0.000056
- Zhang H, de Vries RP, Tzarum N, et al. A human-infecting H10N8 influenza virus retains a strong preference for avian-type receptors. Cell Host Microbe. 2015;17:377–384. doi: 10.1016/j.chom.2015.02.006
- de Vries E, Du W, Guo H, et al. Influenza a virus hemagglutinin–neuraminidase–receptor balance: Preserving virus motility. Trends Microbiol. 2020;28(1):57–67. doi: 10.1016/j.tim.2019.08.010
- Du W, Wolfert MA, Peeters B, et al. Mutation of the second sialic acid-binding site of influenza a virus neuraminidase drives compensatory mutations in hemagglutinin. PLOS Pathog. 2020;16:e1008816. doi: 10.1371/journal.ppat.1008816
- An SH, Son SE, Song JH, et al. Selection of an optimal recombinant egyptian H9N2 avian influenza vaccine strain for poultry with high antigenicity and safety. Vaccines (Basel). 2022;10(2):162. doi: 10.3390/vaccines10020162
- Scheibner D, Salaheldin AH, Bagato O, et al. Phenotypic effects of mutations observed in the neuraminidase of human origin H5N1 influenza a viruses. PLOS Pathog. 2023;19(2):e1011135. doi: 10.1371/journal.ppat.1011135

