ABSTRACT
Clade 2.3.4.4 H5N6 avian influenza virus (AIV) has been predominant in poultry in China, and the circulating haemagglutinin (HA) gene has changed from clade 2.3.4.4h to clade 2.3.4.4b in recent years. In 2021, we isolated four H5N6 viruses from ducks during the routine surveillance of AIV in China. The whole-genome sequencing results demonstrated that the four isolates all belonged to the currently prevalent clade 2.3.4.4b but had different internal gene constellations, which could be divided into G1 and G2 genotypes. Specifically, G1 possessed H9-like PB2 and PB1 genes on the H5-like genetic backbone while G2 owned an H3-like PB1 gene and the H5-like remaining internal genes. By determining the characteristics of H5N6 viruses, including growth performance on different cells, plaque-formation ability, virus attachment ability, and pathogenicity and transmission in different animal models, we found that G1 strains were more conducive to replication in mammalian cells (MDCK and A549) and BALB/c mice than G2 strains. However, G2 strains were more advantageously replicated in avian cells (CEF and DF-1) and slightly more transmissible in waterfowls (mallards) than G1 strains. This study enriched the epidemiological data of H5 subtype AIV to further understand its dynamic evolution, and laid the foundation for further research on the mechanism of low pathogenic AIV internal genes in generating novel H5 subtype reassortants.
Introduction
The highly pathogenic avian influenza virus (HPAIV) H5N1 was first isolated from geese in Guangdong Province in 1996, with the prototype strain A/Goose/Guangdong/1/96. Since then, the Gs/GD-96-like strain has been prevalent in many parts of China and has rapidly spread to Africa, Europe, and other countries [Citation1]. H5 avian influenza virus (AIV) has evolved into numerous genotypes with different neuraminidase (NA) subtypes and ten phylogenetic clades (clades 0 to 9) [Citation2].
HPAIV H5N6 in China was initially reported in 2014, and has spread rapidly across the country within a few years [Citation3,Citation4]. In particular, clade 2.3.4.4 H5N6 is widely prevalent in China, which has globally evolved into eight groups (clade 2.3.4.4a-h) [Citation5]. In 2020, the predominate haemagglutinin (HA) clade of circulating H5N6 viruses in China and other Southeast Asian countries was 2.3.4.4 h, but the prevalent level of clade 2.3.4.4 h H5N6 viruses gradually declined at the end of the year [Citation6–8]. Subsequently, H5N6 viruses of clade 2.3.4.4b emerged and caused several human infections in China [Citation9]. To date, the preponderant HA genes of the H5N6 subtype HPAIV in China have changed from clade 2.3.4.4 h to clade 2.3.4.4b.
As for clade 2.3.4.4b, the early prevailing HPAIV subtype was H5N8 that was first isolated from domestic ducks in China [Citation10,Citation11]. The clade 2.3.4.4b HA gene had further reassorted with NA subtypes N5 and N6 in Europe [Citation12]. However, in late 2020, clade 2.3.4.4b H5N8 from wild birds revived in China [Citation13]. Furthermore, the HA gene of these H5N8 viruses was further reassorted with the then-current H5N6 viruses to generate clade 2.3.4.4b H5N6 variants [Citation5,Citation14]. These H5N6 viruses have been reported to have preferable host tropism for waterfowls [Citation15].
H5N6 AIV infect not only poultry and wild birds, but also humans. As of March 9, 2023, there were 84 cases of H5N6 infection in humans, 33 of which were deaths [Citation16]. Therefore, HPAIV H5N6 poses a serious threat to public health. In this study, four H5N6 viruses bearing circulating clade 2.3.4.4b HA genes but different internal gene compositions were isolated from apparently healthy domestic ducks in live poultry markets (LPMs) in eastern China during routine surveillance of AIV. The virulence and transmission characteristics of H5N6 viruses in BALB/c mice and mallard ducks were investigated.
Materials and methods
Ethic statements
All experiments involving H5 HPAIV were executed in the animal biosafety level 3 facility at Yangzhou University in strict accordance with the recommendations of the institutional biosafety manual. All experimental animal research operations complied with the welfare standards of laboratory animals and were approved by the Jiangsu Provincial Committee for Laboratory Animal Control (Permission number: SYXK-SU-2021–0027, SYXK-SU-2022–0044).
Viruses
Four H5N6 viruses of A/Duck/Shandong/SD0261/2021 (SD0261 for short), A/Duck/Shandong/SD0263/2021 (SD0263 for short), A/Duck/Sichuan/SC5698/2021 (SC5698 for short), and A/Duck/Sichuan/SC4822/2021 (SC4822 for short) were isolated from the throat and cloacal swabs of apparently healthy ducks in LPMs. Viruses were plaque-purified on Madin-Darby canine kidney (MDCK) cells and harvested from 10-day-old specific-pathogen-free (SPF) embryonated eggs by collecting allantoic fluids. The prepared virus stocks were aliquoted and stored at −70℃.
Virus identification and phylogenetic analysis
Viral RNA was extracted from the allantoic fluid using a Genenode RNA extraction kit according to the instructions. cDNA was synthesized by reverse transcription using the primer Uni12 (5’-AGCAAAAGCAGG-3’). The whole genome segments were amplified by PCR using primers described by Hoffmann et al. [Citation17]. After electrophoresis on a 1% agarose gel, the target bands were cut and sent to the sequencing company. The sequences were edited using DNASTAR and DNAMAN software, and the corresponding reference sequences were downloaded from the National Center for Biotechnology Information (NCBI) and Global Initiative on Sharing All Influenza Data (GISAID) databases. For the construction of phylogenetic trees of each gene segment for the four H5N6 viruses, MEGA 11.0 with the distance-based neighbour-joining method was applied.
Viral growth kinetics in different cells
Four H5N6 viruses were measured with the 50% tissue culture infective dose (TCID50) on MDCK cells according to the Reed-Muench method [Citation18]. We then determined viral growth curves in MDCK, adenocarcinoma alveolar basal epithelial (A549), and chicken embryo fibroblast (CEF) cells, respectively. Briefly, each kind of monolayer cells were infected with the viruses at the multiplicity of infection (MOI) of 0.001 in serum-free Dulbecco’s modified Eagle medium (DMEM). The supernatants were collected at 12, 24, 36, 48, 60, and 72 h post-infection (hpi) and titrated in MDCK cells as expressed by TCID50.
Viruses plaque-formation ability in different cells
Four H5N6 viruses were evaluated for plaque-formation assay in MDCK and DF-1 cells at the MOI of 0.001. After virus adsorption for 1.5 h, the infected cells were covered with a mixture of equal volumes of 2× concentrated DMEM (containing 4% foetal bovine serum) and 1.6% agar, and cultured for 60 h. Next, 4% paraformaldehyde fixation and crystal violet staining were used to observe the plaques. We used Nano Measurer 1.2 to measure the diameter of virus plaques.
Attachment capacity of virus in different cells
According to the references [Citation19,Citation20] with slight modifications, CEF or A549 cells were seeded in 6-well plates (approximately 1 × 106.0 cells/per well) for adherent culture for 12 h. After infection with 5 MOI viruses and then incubated at 37℃ for 1 h, the cells were washed trice with PBS. Subsequently, the infected cells were incubated with an anti-HA2 rabbit monoclonal antibody (SinoBiological, Cat: 86001-RM02) at room temperature for 2 h. Following three times of washing with PBST (PBS with 0.05% Tween-20), the cells were treated with fluorescein isothiocyanate (FITC)-labelled goat anti-rabbit IgG at 37℃ in dark for 1 h. Then, the tested cells, after trice washing with PBST, were digested from the 6-well plates to prepare cell suspension for the flow cytometry assay.
Replication and virulence of H5N6 viruses in mice
First, the 50% egg infection dose (EID50) values of the four H5N6 viruses were measured in 10-day-old SPF chicken eggs according to the Reed-Muench method. Subsequently, groups of eleven six-week-old female BALB/c mice were infected intranasally with the four viruses of 106.0 EID50 in 50 μL. In addition, five mice mock-inoculated with PBS were regarded as a control group. Organs, including the lung and brain, were collected at 3 and 5 days post-infection (dpi) for virus titration in SPF chicken eggs as expressed by EID50 (flow chart in Supplementary Figure S1). In addition, lung tissues were fixed with 10% formalin solution and sliced for haematoxylin-eosin (HE) staining and histopathological analysis. Body weight and clinical symptoms of infected mice were recorded daily until 14 dpi to draw the curves of weight changes and survival rates for assessing viral pathogenicity in mice. If the mice lost more than 25% of their initial body weight, they were humanely euthanized and considered dead.
Pathogenicity and transmissibility of H5N6 viruses in mallard ducks
Three-week-old mallard ducks were randomly divided into two groups of 14 ducks per group and inoculated intranasally with the two viruses (SD0263 and SC4822) of 106.0 EID50 in 200 μL. Specifically for the 14 challenged ducks in either virus group, 5 of which were monitored morbidity and mortality, as well as virus shedding from the throat and cloaca. The other 9 ducks were used for evaluating virus replication in different tissues, with 3 ducks per day being randomly killed on 1, 3, 5 dpi to collect tissue samples involving the liver, spleen, lung, kidney, brain, trachea, pancreas, intestine, and cloaca for virus titration in SPF chicken eggs, as expressed by EID50. If some of the 9 ducks per group died on 2 or 4 dpi, their tested data were calculated into the following day for statistical analysis.
For further evaluating the transmissibility of SD0263 and SC4822 viruses in mallard ducks, at 24 hours post challenge, another five naïve SPF ducks were fed together with either of the inoculated groups as a contact group. For all these contact ducks plus the above mentioned five challenged ducks in both viral groups, swabs were collected from the throat and cloaca at 3, 5, 7, 9, 11, and 13 dpi to detect virus shedding through inoculation into SPF chicken eggs. Additionally, clinical symptoms were monitored daily to evaluate the pathogenicity and transmissibility of the two viruses in ducks. At 14 dpi, the serum of surviving ducks was collected for determination of seroconversion by haemagglutination inhibition (HI) assay (flow chart in Supplementary Figure S2).
Results
Genetic evolution analysis of the four H5N6 virus isolates
We conducted genome sequencing of the four H5N6 AIV strains isolated in early 2021 (GenBank accession numbers: ON862693-ON862724). Sequencing results showed that the open reading frame (ORF) of the same gene segments were highly homologous among the four H5N6 viruses. However, they could be further divided into two genotypes based on their different internal gene compositions. In particular, SD0261 and SD0263 both possessed H9N2-like PB2 and PB1 genes plus H5-like PA, HA, NP, NA, M, and NS genes, and were defined as G1 genotype viruses. In contrast, SC5698 and SC4822 both owned H3-like PB1 genes while the other seven genes were all H5-like, and were designated as G2 genotype viruses ().
Figure 1. The strains with the highest similarity to the four H5N6 viruses through BLAST search against the NCBI GenBank database. Eight lines inside the ellipse represent the eight gene segments (PB2, PB1, PA, HA, NP, NA, M, NS from top to bottom). Different colours represent different sources of gene segments, with H9-like in green, H5-like in red, H6-like in blue and H3-like in grey. The peripheral strain indicated the one with the highest nucleotide identity of corresponding gene in the NCBI GenBank database.
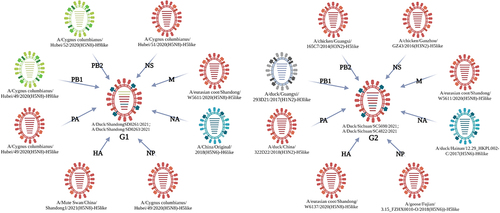
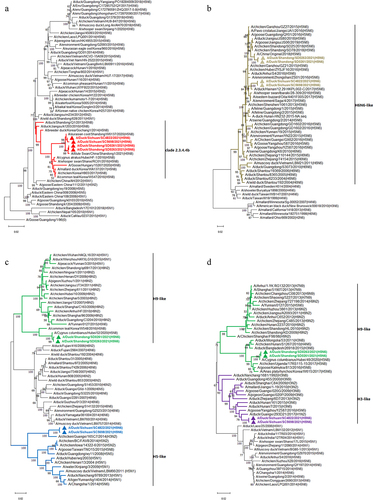
The ORF nucleotide identities of HA genes among the four H5N6 viruses were 99.05%-100.00%, in which SD0261 and SD0263 were 100% consistent. Although the G1 and G2 strains shared extremely similarity at the amino acid level of HA gene, they differed obviously at site 192 (H3 numbering) in the receptor binding region, with the former of M and the latter of I. The nucleotide similarities of the four NA genes were 93.13%-99.86%, with several different amino acid sites between G1 and G2 strains, such as 26 T, 48–50 LET, 63–65 IKV, 70 G, 100 G, 216 H, 251 I, 275 G, 294 I, 299 M, 311 T, and 375 N in G1 strains, while 26 I, 48–50 PNM, 63–65 TIM, 70 E, 100 D, 216 Q, 251 K, 275 R, 294 T, 299 R, 311 K, and 375 D in G2 strains. In terms of the internal genes, the nucleotide sequence divergence of the four analytical strains were 91.85%-99.93% for PB2, 94.07%-100.00% for PB1, 91.13%-100.00% for PA, 93.89%-100.00% for NP, 99.59%-100.00% for M, and 89.11%-100.00% for NS, respectively. We also noticed that the two G1 genotype strains shared 100% identical coding regions for PB1, PA, HA, NP, NA, and M genes, but differed in their PB2 and NS genes. In addition, the two G2 genotype strains had the same nucleotide coding sequences as the NS gene. As revealed by the phylogenetic trees of the gene segments of the four H5N6 viruses (), the HA genes uniquely belonged to clade 2.3.4.4b (), whereas the NA genes clustered into the H6N6 virus lineage of Eurasian avian origin (). For internal genes, the PB2 genes dispersed in two branches of H9-like and H5-like, while their PB1 genes diverged into H9-like and H3-like (). The remaining PA, NP, M, and NS genes were derived from the H5-like AIV (Supplementary Figure S3).
Figure 2. Phylogenetic trees of the four H5N6 subtype viruses. (a) phylogenetic tree of HA gene, the red branches indicate clade 2.3.4.4b and the red triangles label the four H5N6 viruses in this study. (b) phylogenetic tree of NA gene, the brown branches indicate H6-like strains and the brown triangles label the four H5N6 viruses in this study. (c) phylogenetic tree of PB2 gene, the green branches indicate H9-like strains while the blue branches indicate H5-like strains, and the triangles label the four H5N6 viruses in this study. (d) phylogenetic tree of PB1 gene, the green branches indicate H9-like strains while the purple branches indicate H3-like strains, and the triangles label the four H5N6 viruses in this study. All those phylogenetic trees were constructed using MEGA 11.0 software, with the neighbour-joining method and 1000 replicates of bootstrap analysis.
The HA cleavage sites of four H5N6 isolates carried multiple consecutive basic amino acids, which is consistent with the molecular characteristics of HPAIV. Each virus possessed 226Q and 228 G at the HA receptor-binding sites, indicating a stronger affinity for avian-type α-2,3 sialic acid receptors. However, site substitutions of S137A, S158N, and T160A in HA protein also suggest the potential for binding to human-type α-2,6 sialic acid receptors [Citation21]. An 11-amino-acid deletion at positions 58–68 in the stem of NA protein, which may relate to increased viral adaptation from waterfowl to terrestrial poultry and pathogenicity to mammals, was also noted for the four H5N6 viruses [Citation22]. Key amino acid mutations such as A274T, E627K, and D701N of PB2 protein that can improve polymerase activity and increase adaptability to mammals, or H99Y and I368V of PB1 protein, which can enhance virus transmission among ferrets [Citation23,Citation24], were not found in the H5N6 isolates. However, 66S of the PB1-F2 protein, which has been reported to significantly strengthen viral pathogenicity in mice [Citation25] were present in all four H5N6 viruses.
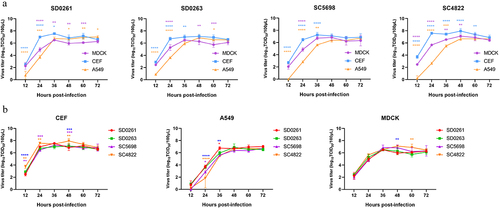
Growth kinetics of the H5N6 viruses in three cell types
The determined TCID50/100 μL values of SD0261, SD0263, SC5698, and SC4822 strains in MDCK cells were 107.67, 107.67, 107.67, and 107.5, respectively. At an inoculation dosage of 0.001 MOI, the growth kinetics of the four viruses were measured in MDCK, CEF, and A549 cells. As shown in , the viral replication performance of all tested H5N6 viruses in avian cells (CEF) was significantly higher than that in mammalian cells (MDCK and A549) during the first 36 h, and the most different time point was at 24 hpi. showed that especially in CEF cells, the replication titres of SC4822 were significantly higher than those of the other G1 strain SC5698 at 12, 24, and 48 hpi. However, after 60 hpi, viral growth in CEF cells was inferior to that in A549 cells (). In addition, only G1 genotype viruses of SD0261 and SD0263, but not the two G2 genotype viruses, could detect virus titres in A549 cells at 12 hpi (). G1 strains were more replicated than G2 strains during 24–36 hpi (). In MDCK cells, the proliferation efficiency of G1 strains peaked at 36 hpi and then decreased within 12 h while G2 strains peaked at a later time point of 48 hpi but with relatively higher virus titres ().
Figure 3. Growth kinetics of the four H5N6 viruses in three cell types of cells. (a) growth curves of the same H5N6 strain in three different cells. (b) growth curves of the four H5N6 viruses in the same cell type. CEF, MDCK, and A549 cells were each infected with the four H5N6 viruses at the MOI of 0.001. After virus adsorption of 1.5 h, infected cells were cultured in serum-free DMEM for 12–72 h. Virus titres were measured at 12, 24, 36, 48, 60, and 72 h post-infection (hpi), and expressed as mean (n = 3) ± standard deviation. In panel a, purple *means MDCK versus CEF, yellow *means MDCK versus A549, and blue *means CEF versus A549. In panel b, blue *means SD0261 versus SC4822, purple *means SC5698 versus SC4822, yellow *means SD0263 versus SC4822, red *means SD0261 versus SC5698. Different numbers of *denote different p-value thresholds (*<0.05, **<0.01, ***<0.001, ****< 0.0001).
Plaque-formation ability of the H5N6 viruses in different cells
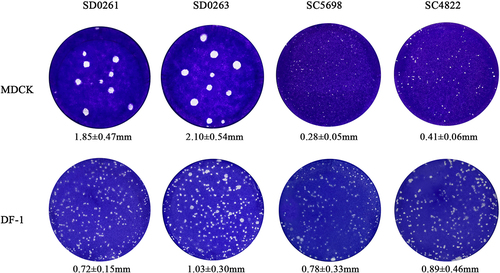
We further evaluated the plaque-formation ability of the four H5N6 viruses in mammalian MDCK and avian DF-1 cells at the same dose (0.001 MOI). It was apparent that the plaques formed by G1 strains of SD0261 and SD0263 in MDCK cells were significantly larger than those in DF-1 cells. In contrast, the plaque diameters of G2 strains of SC5698 and SC4822 in DF-1 cells were significantly larger than those in MDCK cells (). The above results showed that genotypes G1 and G2 H5N6 subtype AIV of clade 2.3.4.4b were more conducive to replication in mammalian and avian cells, respectively.
Figure 4. Plaque-formation capacity of the four H5N6 viruses in different cells. MDCK and DF-1 cells were infected with the MOI of 0.001 for 1.5 h and covered with the mixture of 2× concentrated DMEM (containing 4% foetal bovine serum) and 1.6% agar. After 60 h, infected cells were fixed with 4% paraformaldehyde for 15 min and then stained with crystal violet staining solution for 1 h. Plaques were randomly selected for measurement of the diameter size. Data were expressed as the mean ± standard deviation.
Attachment capacity of the H5N6 viruses in different species cells
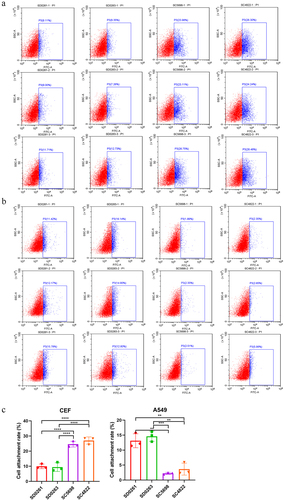
As adsorption is the first step for viral infection of host cells, we measured the binding rate of the four H5N6 viruses in CEF and A549 cells using flow cytometry to evaluate their infectivity differences in vitro. The results showed that the attachment rates of G2 SC5698 and SC4822 strains on CEF cells were significantly higher than those of G1 SD0261 and SD0263 strains (). Specifically, G2 SC4822 possessed the highest attachment efficiency, which was consistent with previous growth curve results in CEF cells (). On the contrary in A549 cells, the attachment capacities of G1 SD0261 and SD0263 strains were stronger than G2 SC5698 and SC4822 strains (). This attachment assay again supported that G1 strains were more favourable to mammalian cells, whereas G2 strains were more conducive to avian cells.
Figure 5. The attachment capacity of the four H5N6 viruses in CEF and A549 cells. (a) viral attachment assay in CEF cells. (b) viral attachment assay in A549 cells. (c) data were expressed as mean viral attachment capacity (n = 3) ± standard deviation. CEF or A549 cells were seeded in 6-well plates (approximately 1 × 106.0 cells/per well) and infected with 5 MOI viruses at 37℃ for 1 h. After incubation with an anti-HA2 rabbit monoclonal antibody (SinoBiological, Cat: 86001-RM02) at room temperature for 2 h, the cells were washed with PBST (PBS with 0.05% Tween-20) and treated with fluorescein isothiocyanate (FITC)-labelled goat anti-rabbit IgG at 37℃ in dark for 1 h. Then, the tested cells were digested from the 6-well plates to prepare cell suspension for the flow cytometry assay.
Replication and virulence of the H5N6 viruses in mice
The four H5N6 strains each showed vigorous replication performance in SPF chicken eggs, with the EID50/100 μL values of 108.167 for SD0261, 108.5 for SD0263, 108.625 for SC5698, and 108.625 for SC4822, respectively. We then challenged BALB/c mice with a dose of 106.0 EID50 in 50 μL for every tested H5N6 virus. As shown in , SD0261 caused the death of mice as early as 4 dpi, and both G1 strains induced complete mortality at 6 dpi. However, the G2 strain resulted in relatively milder body weight loss, and the survival rate of inoculated mice reached 60%. Virus titration in tissues showed all four H5N6 strains replicated efficiently in the lungs without prior adaptation at 3 and 5 dpi (). For virus replication in the brain, only SD0261 and SD0263 showed virus titres at 3dpi. Although 1/3 of the sampled mice infected with G2 SC5698 and SC4822 strains were virus-positive at 5 dpi, their viral titres were significantly lower than those of the two G1 strains.
Figure 6. Pathogenicity and replication of the four H5N6 viruses in mice. (a) body weight changes and survival curves of mice infected with the four H5N6 viruses. Four groups of five six-week-old female BALB/c mice were inoculated intranasally with 106.0 EID50 virus in 50 μL volume, and another group of five mice was mock-infected with PBS to serve as a control. The weight changes and survival of mice were recorded daily for 14 days. (b) replication of the four H5N6 viruses in the lung and brain of infected mice. Virus replication in organs was assessed by mean viral titres (n = 3) ± standard deviation. Different numbers of *denote different p-value thresholds (*<0.05, **<0.01, ***<0.001, ****< 0.0001). (c) HE staining of sectioned lungs from mice infected with the four H5N6 viruses on 3 and 5 days post-inoculation (dpi). The histological sections were observed and photographed at 200 × magnification.
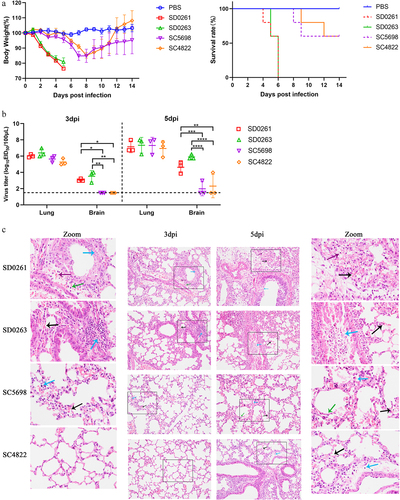
As shown in , HE staining of the lungs at 3 dpi revealed that the normal tissue structure was preserved in the SC4822-infected mice, while only a few inflammatory cells (black arrows) and macrophages (blue arrows) were observed in the SC5698-infected mice. Compared to the two G2 genotype H5N6 viruses, the G1 strains induced relatively more severe historical changes. Specifically, thickened alveolar walls, exfoliated epithelial cells (blue arrow), lymphocytes (green arrow), and minor haemorrhage (purple arrow) were observed in representative pathological sections from SD0261 group. Similarly, the section from SD0263 group also revealed minor haemorrhage (black arrow) and a few lymphocytes (blue arrow) in the lungs. At 5 dpi, increased numbers of lymphocytes (purple arrows) were found in SD0261-infected mouse lungs, with karyopyknosis of bronchial epithelial cells (blue arrow) and karyorrhexis of alveolar epithelial cells (green arrow). For SD0263-infected mice, inflammatory cells (black arrow) and abruption, necrosis of bronchial epithelial cells (blue arrow), were observed in the lung. The G2 strains of SC5698 and SC4822 both displayed focal thickening of the alveolar walls with slight haemorrhage (black arrow). In addition, lymphocyte infiltration (blue arrow) was noted in SC4822 group, whereas a small number of pulmonary macrophages (blue arrow) and enlarged alveolar walls (green arrow) were observed in SC5698 group. Overall, the above pathogenicity tests in mice demonstrated that the G1 strains were more virulent and replicable in mammals than the G2 strains ().
Transmissibility and pathogenicity of the H5N6 viruses in mallard ducks
The G1 genotype strain SD0263 and G2 genotype strain SC4822 were selected for animal experiments in mallard ducks. Apart from the nine ducks per inoculation group that were humanely killed for tissue collection, the remaining five inoculated ducks as well as the five contact ducks in either virus group were monitored for disease and mortality until 14 dpi. Although the two H5N6 viruses shared identical survival rates of ducks (20%) at the final observation time points in both inoculation and contact groups, SC4822 induced relatively severe death at 4–6 dpi than SD0263 in the inoculated group, but more delayed death at 6 and 7 dpi than SD0263 in the contact group (). Except for the pancreas and intestine, all the other collected organs at 1 dpi were 100% (3/3) virus-positive with the highest viral titres in the lung. At 3 and 5 dpi, generally increased virus titres were detected in all sampled organs, including the lung, liver, spleen, kidney, brain, trachea, pancreas, intestine, and cloaca (). Regarding the level of virus shedding in the inoculated ducks, neither SD0263 nor SC4822 virus could be recovered from throat swabs at 7 dpi. However, SC4822 extended cloacal shedding to 9 dpi, whereas SD0263 only terminated at 7 dpi. In contact ducks, positive throat shedding lasted until 9 dpi for SD0263 virus, whereas SC48223 virus could only be detected at 3 and 5 dpi. However, virus recovery from cloacal swabs was negative for SD0263 after 5 dpi, while SC4822 could still be detected at 7 dpi (). At 14 dpi, only one duck survived per virus per group, and all four ducks seroconverted (). Therefore, although SD0263 and SC4822 possessed comparable pathogenicity in mallard ducks, the G2 strain SC4822 was slightly more transmissible due to its long-lasting cloacal shedding in both inoculated and contact groups.
Figure 7. Pathogenicity and transmission of the four H5N6 viruses in mallard ducks. (a) survival curves of inoculated and contact ducks in either virus group. (b) replication of the four H5N6 viruses in different tissues of challenged ducks. Two groups of fourteen three-week-old SPF mallard ducks were inoculated intranasally with SD0263 (G1 genotype) and SC4822 (G2 genotype) viruses of 106.0 EID50 in 200 μL volume per duck. At 24 h post-inoculation (hpi), two groups of five naïve ducks were fed together with either inoculated group to serve as direct-contact individuals. For inoculated ducks in either group, five were daily monitored disease and death until 14 days post-inoculation (dpi) while the other nine ducks were humanely killed on 1, 3, and 5 days (three ducks per day) for evaluation of virus replication in different tissues. In addition, mortality of all the contact ducks was also recorded until 14 dpi. Virus replication in different tissues of inoculated ducks was evaluated by mean virus titres ± standard deviation (n = 3).
Table 1. Virus shedding in throat and cloaca of inoculated and contact mallard ducks.
Discussion
Since clade 2.3.4.4b H5 AIV revived at the end of 2020, the HA gene has been reassorted with a variety of NA subtypes to generate novel reassortants, including H5N1, H5N2, H5N3, H5N4, H5N5, H5N6, and H5N8 viruses. H5NX variants have spread to many countries in Europe, Asia, and Africa, causing serious threats to both the poultry industry and public health [Citation26,Citation27]. On the other hand, H9N2 has long been considered one of the hightest isolated LPAIV subtypes in live poultry markets. Its endemicity in China together with the prevalence of H5NX would inevitably promote the occurrence of gene reassortment. As previously reported by Hao et al., H9N2 internal gene cassette involved in H5 reassortants could significantly weaken the pathogenicity in chickens and mice [Citation28]. Subsequently, Liu et al. isolated natural reassortant H5N6 viruses containing all internal genes from H9N2, and revealed their high pathogenicity in chickens, low pathogenicity in mice, and asymptomatic infections in ducks [Citation29]. In addition, H9N2 has frequently donated part or whole set of internal genes to generate novel human-infecting influenza reassortants, similar to the recently reported H3N8 variants [Citation30]. In addition, these studies indicated that H9N2 internal genes might contribute to viral dissemination and adaptation. Other than H9N2, LPAIV including H3, H4, and H6 subtypes, were found to participate in the genetic reassortment of H5N6 viruses [Citation21,Citation31,Citation32]. Furthermore, several human H5N6 isolates possessed internal genes derived from LPAIV. Therefore, the involvement of LPAIV internal genes in novel H5N6 reassortants should be paid attention to.
In this study, four H5N6 AIV strains were isolated from ducks in early 2021. Phylogenetic analysis showed that they share different lineages of PB2 and PB1 genes, which caused division into two genotypes. G1 strains of SD0261 and SD0263 owned H9-like PB2 and PB1 genes while G2 strains of SC4822 and SC5698 possessed H5-like PB2 and H3-like PB1 genes, with all the other six gene segments of H5-like. We then compared the viral biological properties of the two genotypes in vitro and in vivo. Through flow cytometry, we found that the attachment capacity of G2 strains was significantly higher than that of G1 strains in CEF but lower than that of G1 strains in A549 cells (). Despite that the G1 and G2 strains shared extremely similar HA genes at the amino acid level, they were different at site 192 in the receptor binding region. And this discrepancy at site 192 might be a key factor affecting the viral attachment ability, which required further experimental verification. On the other hand, it is more and more recognized that the HA and NA proteins share functional balance [Citation33], thus the difference of NA genes may also have the potential to affect receptor binding through interfere with HA-NA balance. It was reasonable to speculate that the several genotype-specific different sites in the NA gene might also be a possible contributing factor to impact the viral attachment capacity of G1 and G2 strains. Similarly, the viral growth curves showed that the early replication capacity of G1 was significantly higher than that of G2 strains in A549 cells, and the maximum replication of G1 peaked 12 h earlier than G2 in MDCK cells (). In addition, the plaque-formation assay revealed that G1 strains produced obviously larger plaque diameters than that of G2 strains in A549 cells (). All these above results consistently indicate that G1 viruses containing H9-like PB2 and PB1 genes may replicate more advantageously in mammalian cells than G2 viruses.
Further pathogenicity tests in mice showed that the G1 strains were more virulent to mice than G2 viruses, with G1 causing a mortality rate of up to 100% within 6 dpi, and G2 maintained a survival rate of 60% at 106.0 EID50 challenge conditions (). As G1 and G2 viruses mainly differed in PB2 and PB1 lineages from the perspective of gene constellations, we inferred that the H9-like PB2 and PB1 genes might be responsible for pathogenicity to mammals. Several novel AIV reassortants, including H3N8, H5N1, H5N6, H7N9, H10N3, and H10N8, which harbour H9-like internal genes, are zoonotic and even harmful to humans [Citation34–36]. Thus, the specific roles of these H9-like PB2 and PB1 genes and their related mechanisms require further exploration.
Waterfowls, especially ducks, constitute the primary natural reservoir for AIV and rarely display obvious clinical symptoms when infected [Citation37]. However, the global-circulating clade 2.3.4.4 H5NX viruses have caused severe damage to wild waterfowls [Citation38,Citation39]. Several reports have also indicated that wild bird H5NX viruses can replicate efficiently in ducks and geese with either high or mild pathogenicity in these birds [Citation23,Citation40]. To investigate the status of our H5N6 viruses in waterfowl, we chose G1 SD0263 and G2 SC4822 strains to conduct pathogenicity assay in mallard ducks. The results showed that both H5N6 viruses propagated effectively in nearly all collected duck tissues (). SD0263 and SC4822 both induced the same survival rate (20%) in both the inoculated and contact groups (). But the virus dose of 106.0 EID50 in 200 μL failed to maintain all the challenged ducks alive during the test period. Less than expected ducks were left for comparison since 4 dpi. We noticed that G2 SC4822 caused relatively earlier death than G1 SD0263 in the inoculated groups. The slightly higher pathogenicity of G2 SC4822 in poultry was consistent with the previous finding that G2 strains adsorbed () more efficiently in avian cells. Although extended throat shedding of G1 SD0263 was observed in the contact groups, G2 SC4822 consistently caused prolonged cloacal shedding in both the inoculated and contact groups (), indicating that G1 H5N6 strains with H9-like PB2 and PB1 were less transmissible in waterfowl than G2 strains containing H5-like PB2 and H3-like PB1 genes.
The RNA polymerase complex of the influenza virus is a heterotrimeric structure composed of PB2, PB1, and PA proteins, which mainly participate in the replication and transcription process after virus infection and entry into host cells [Citation41]. The PB1 protein is located in the centre of the complex, with its N-terminal and C-terminal interacting with PA and PB2 proteins, respectively [Citation42]. As reported, PB1 protein can affect viral replication through post-translational modifications, such as phosphorylation and SUMOylation [Citation43–45]. Specifically, phosphorylation may be related to nuclear import and localization, whereas SUMOylational modification can promote the binding of viral RNA to proteins and affect viral pathogenicity and transmission [Citation43–45]. In addition to PB1 protein, AIV gene segment 2 encodes PB1-F2 and N40 proteins through ribosomal leaky scanning and reinitiation. The PB1-F2 protein can affect viral polymerase activity and viral replication efficiency, regulate innate immune responses, and induce cell apoptosis [Citation46–48]. Moreover, the PB1 N40 protein has also been reported to influence polymerase activity and viral replication, and may be related to the expression balance of PB1 and PB1-F2 proteins [Citation49]. For PB2 protein, the key amino acid substitution R124A in the N-terminal domain was shown to play an important role in promoter binding during viral RNA (vRNA) synthesis, thus significantly reducing the synthesis of vRNA, mRNA, and complementary RNA (cRNA) to affect viral replication [Citation50]. Structurally, the cap-binding domain of PB2 protein can seize the 5’-terminal primers of host mRNA, altering polymerase activity and transcriptional synthesis [Citation51]. The nuclear localization signal (NLS) domain can bind import-α into the nucleus to initiate transcription and replication [Citation41]. Furthermore, several site mutations including A588V, E627K, and D701N adjacent to the C-terminus of PB2 protein were essentially involved in viral replication and pathogenicity in mammals [Citation52]. In this study, we found that G1 and G2 strains bearing PB2 and PB1 genes from different virus lineages resulted in significant variations in biological properties in vitro and in vivo. Whether such divergence of PB2 and PB1 genes from H9-like, H5-like, or H3-like affected polymerase activity, viral replication and transcription, as well as the relevant mechanisms remains to be further studied.
In summary, four clade 2.3.4.4b H5N6 subtype AIV strains were genetically and phenotypically characterized in the present study. The results showed that internal genes from H9 and H3-like LPAIV were continuously reassorted with the circulating clade 2.3.4.4b H5N6 viruses, and these novel reassortants possessed efficient transmissibility among waterfowls and high pathogenicity to mammals. Therefore, to avoid the spread and dissemination of the H5 subtype HPAIV in China, sustained monitoring is indispensable to master the evolution dynamics in a timely manner, apart from the enforcement of a stamping-out plus compulsory vaccination policy. Additionally, the functional role of LPAIV-like internal genes in the generation of novel influenza reassortants also deserves more attention to better understand viral pathogenesis and zoonotic transmission.
Author contributions
MG, XFL, and WCZ conceived and designed the study. WCZ, XL, XYZ, ZWQ, JJ, and YL performed experiments. RYG, XQW, JH, XWL, SLH, XAJ, and DXP discussed and analysed the data. WCZ and MG wrote the manuscript.
Supplemental Material
Download Zip (3.2 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data Availability statement
The data supporting the findings of this study are available within the article and its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2023.2250065.
Additional information
Funding
References
- Nunez IA, Ross TM. A review of H5Nx avian influenza viruses. Ther Adv Vaccines Immunother. 2019;7:2515135518821625. doi: 10.1177/2515135518821625
- Revised and updated nomenclature for highly pathogenic avian influenza a (H5N1) viruses . Influenza and other respiratory viruses. Influenza Other Respir Viruses. 2014;8(3):384–14. doi: 10.1111/irv.12230
- Zhang R, Chen T, Ou X, et al. Clinical, epidemiological and virological characteristics of the first detected human case of avian influenza A(H5N6) virus. Infect Genet Evol. 2016;40:236–242. doi: 10.1016/j.meegid.2016.03.010
- Bi Y, Chen Q, Wang Q, et al. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20(6):810–821. doi: 10.1016/j.chom.2016.10.022
- Zhang Q, Mei X, Zhang C, et al. Novel reassortant 2.3.4.4B H5N6 highly pathogenic avian influenza viruses circulating among wild, domestic birds in Xinjiang, Northwest China. J Vet Sci. 2021;22(4):e43. doi: 10.4142/jvs.2021.22.e43
- Li Y, Li M, Li Y, et al. Outbreaks of highly pathogenic avian influenza (H5N6) virus Subclade 2.3.4.4h in Swans, Xinjiang, Western China, 2020. Emerg Infect Dis. 2020;26(12):2956–2960. doi: 10.3201/eid2612.201201
- Turner JCM, Barman S, Feeroz MM, et al. Highly pathogenic avian influenza A(H5N6) virus clade 2.3.4.4h in wild birds and live poultry markets, Bangladesh. Emerg Infect Dis. 2021;27(9):2492–2494. doi: 10.3201/eid2709.210819
- Jeong S, Otgontogtokh N, Lee DH, et al. Highly pathogenic avian influenza clade 2.3.4.4 subtype H5N6 viruses isolated from wild Whooper Swans, Mongolia, 2020. Emerg Infect Dis. 2021;27(4):1181–1183. doi: 10.3201/eid2704.203859
- Gu W, Shi J, Cui P, et al. Novel H5N6 reassortants bearing the clade 2.3.4.4b HA gene of H5N8 virus have been detected in poultry and caused multiple human infections in China. Emerg Microbes Infect. 2022;11(1):1174–1185. doi: 10.1080/22221751.2022.2063076
- Shi W, Gao GF. Emerging H5N8 avian influenza viruses. Science. 2021;372(6544):784–786. doi: 10.1126/science.abg6302
- Zhao K, Gu M, Zhong L, et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet Microbiol. 2013;163(3–4):351–357. doi: 10.1016/j.vetmic.2012.12.025
- Verhagen JH, Fouchier RAM, Lewis N. Highly pathogenic avian influenza viruses at the wild–domestic bird interface in Europe: future directions for Research and surveillance. Viruses. 2021;13(2):212. doi: 10.3390/v13020212
- Li X, Lv X, Li Y, et al. Emergence, prevalence, and evolution of H5N8 avian influenza viruses in central China, 2020. Emerg Microbes Infect. 2022;11(1):73–82. doi: 10.1080/22221751.2021.2011622
- Cui Y, Li Y, Li M, et al. Evolution and extensive reassortment of H5 influenza viruses isolated from wild birds in China over the past decade. Emerg Microbes Infect. 2020;9(1):1793–1803. doi: 10.1080/22221751.2020.1797542
- Seekings AH, Warren CJ, Thomas SS, et al. Highly pathogenic avian influenza virus H5N6 (clade 2.3.4.4b) has a preferable host tropism for waterfowl reflected in its inefficient transmission to terrestrial poultry. Virology. 2021;559:74–85. doi: 10.1016/j.virol.2021.03.010
- WHO. Human infection with avian influenza A(H5) viruses [M/OL]. 2020. https://apps.who.int/iris/bitstream/handle/10665/365675/AI-20230310.pdf?sequence=76&isAllowed=y.
- Hoffmann E, Stech J, Guan Y, et al. Universal primer set for the full-length amplification of all influenza a viruses. Arch Virol. 2001;146(12):2275–2289. doi: 10.1007/s007050170002
- Reed LJ, Muench H. A Simple method of estimating fifty per cent endpoints12. Am J Epidemiol. 1938;27(3):493–497. doi: 10.1093/oxfordjournals.aje.a118408
- Wang Z, Yang H, CHEN Y, et al. A single-amino-acid substitution at position 225 in hemagglutinin alters the transmissibility of Eurasian avian-like H1N1 swine influenza virus in guinea pigs. J Virol. 2017;91(21): doi: 10.1128/JVI.00800-17
- Gao R, Gu M, Shi L, et al. N-linked glycosylation at site 158 of the HA protein of H5N6 highly pathogenic avian influenza virus is important for viral biological properties and host immune responses. Vet Res. 2021;52(1):8. doi: 10.1186/s13567-020-00879-6
- Huang J, Wu S, Wu W, et al. The biological characteristics of novel H5N6 highly pathogenic avian influenza virus and its pathogenesis in ducks. Front Microbiol. 2021;12:628545. doi: 10.3389/fmicb.2021.628545
- Munier S, Larcher T, Cormier-aline F, et al. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol. 2010;84(2):940–952. doi: 10.1128/JVI.01581-09
- Guo F, Li Y, Yu S, et al. Adaptive evolution of human-isolated H5Nx avian influenza a viruses. Front Microbiol. 2019;10:1328. doi: 10.3389/fmicb.2019.01328
- Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541. doi: 10.1126/science.1213362
- Conenello GM, Zamarin D, Perrone LA, et al. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza a viruses contributes to increased virulence. PLoS Pathog. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141
- Lewis NS, Banyard AC, Whittard E, et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg Microbes Infect. 2021;10(1):148–151. doi: 10.1080/22221751.2021.1872355
- Cui P, Shi J, Wang C, et al. Global dissemination of H5N1 influenza viruses bearing the clade 2.3.4.4b HA gene and biologic analysis of the ones detected in China. Emerg Microbes Infect. 2022;11(1):1693–1704. doi: 10.1080/22221751.2022.2088407
- Hao X, Wang J, Hu J, et al. Internal gene Cassette from a genotype S H9N2 avian influenza virus Attenuates the pathogenicity of H5 viruses in chickens and mice. Front Microbiol. 2017;8:1978. doi: 10.3389/fmicb.2017.01978
- Liu K, Gu M, Hu S, et al. Genetic and biological characterization of three poultry-origin H5N6 avian influenza viruses with all internal genes from genotype S H9N2 viruses. Arch Virol. 2018;163(4):947–960. doi: 10.1007/s00705-017-3695-4
- Yang R, Sun H, Gao F, et al. Human infection of avian influenza a H3N8 virus and the viral origins: a descriptive study. The Lancet Microbe. 2022;3(11):e824–e834. doi: 10.1016/S2666-5247(22)00192-6
- Ge Z, Gu M, Cai T, et al. Phylogenetic tracing and biological characterization of a novel clade 2.3.2.1 reassortant of H5N6 subtype avian influenza virus in China. Transbound Emerg Dis. 2021;68(2):730–741. doi: 10.1111/tbed.13736
- Bo H, Zhang Y, Dong J, et al. Distribution and gene characteristics of H3, H4 and H6 subtypes of low pathogenic avian influenza viruses in environment related avian influenza viruses during 2014-2021 in China. Zhonghua Yu Fang Yi Xue Za Zhi. 2022;56(11):1549–1553. doi: 10.3760/cma.j.cn112150-20220810-00803
- De Vries E, Du W, Guo H, et al. Influenza a virus hemagglutinin-neuraminidase-receptor balance: Preserving virus Motility. Trends Microbiol. 2020;28(1):57–67. doi: 10.1016/j.tim.2019.08.010
- Gu M, Xu L, Wang X, et al. Current situation of H9N2 subtype avian influenza in China. Vet Res. 2017;48(1):49. doi: 10.1186/s13567-017-0453-2
- Liu K, Ding P, Pei Y, et al. Emergence of a novel reassortant avian influenza virus (H10N3) in Eastern China with high pathogenicity and respiratory droplet transmissibility to mammals. Sci China Life Sci. 2022;65(5):1024–1035. doi: 10.1007/s11427-020-1981-5
- Tan X, Yan X, Liu Y, et al. A case of human infection by H3N8 influenza virus. Emerg Microbes Infect. 2022;11(1):2214–2217. doi: 10.1080/22221751.2022.2117097
- Scheibner D, Breithaupt A, Luttermann C, et al. Genetic determinants for virulence and transmission of the panzootic avian influenza virus H5N8 clade 2.3.4.4 in Pekin ducks. J Virol. 2022;96(13):e0014922. doi: 10.1128/jvi.00149-22
- Leyson CM, Youk S, Ferreira HL, et al. Multiple gene segments are associated with enhanced virulence of clade 2.3.4.4 H5N8 highly pathogenic avian influenza virus in Mallards. J Virol. 2021;95(18):e0095521. doi: 10.1128/JVI.00955-21
- Wille M, Barr IG. Resurgence of avian influenza virus. Science. 2022;376(6592):459–460. doi: 10.1126/science.abo1232
- Zeng X-Y, He X-W, Meng F, et al. Protective efficacy of an H5/H7 trivalent inactivated vaccine (H5-Re13, H5-Re14, and H7-Re4 strains) in chickens, ducks, and geese against newly detected H5N1, H5N6, H5N8, and H7N9 viruses. J Integr Agr. 2022;21(7):2086–2094. doi: 10.1016/S2095-3119(22)63904-2
- Ling YH, Wang H, Han MQ, et al. Nucleoporin 85 interacts with influenza a virus PB1 and PB2 to promote its replication by facilitating nuclear import of ribonucleoprotein. Front Microbiol. 2022;13:895779. doi: 10.3389/fmicb.2022.895779
- Pflug A, Guilligay D, Reich S, et al. Structure of influenza a polymerase bound to the viral RNA promoter. Nature. 2014;516(7531):355–360. doi: 10.1038/nature14008
- Dawson AR, Wilson GM, Freiberger EC, et al. Phosphorylation controls RNA binding and transcription by the influenza virus polymerase. PLoS Pathog. 2020;16(9):e1008841. doi: 10.1371/journal.ppat.1008841
- Li J, Liang L, Jiang L, et al. Viral RNA-binding ability conferred by SUMOylation at PB1 K612 of influenza a virus is essential for viral pathogenesis and transmission. PLoS Pathog. 2021;17(2):e1009336. doi: 10.1371/journal.ppat.1009336
- Wang G, Zhao Y, Zhou Y, et al. PIAS1-mediated SUMOylation of influenza a virus PB2 restricts viral replication and virulence. PLoS Pathog. 2022;18(4):e1010446. doi: 10.1371/journal.ppat.1010446
- Boravleva E, Treshchalina A, Postnikova Y, et al. Molecular characteristics, receptor specificity, and pathogenicity of avian influenza viruses isolated from wild ducks in Russia. Int J Mol Sci. 2022;23(18):10829. doi: 10.3390/ijms231810829
- Xiao Y, Evseev D, Stevens CA, et al. Influenza PB1-F2 inhibits avian MAVS signaling. Viruses. 2020;12(4):409. doi: 10.3390/v12040409
- Wang Y, Wang J. PB1F2 from influenza a virus regulates the interaction between cytochrome C and cardiolipin. Membranes (Basel). 2022;12(8):795. doi: 10.3390/membranes12080795
- Vasin AV, Temkina OA, Egorov VV, et al. Molecular mechanisms enhancing the proteome of influenza a viruses: an overview of recently discovered proteins. Virus Res. 2014;185:53–63. doi: 10.1016/j.virusres.2014.03.015
- Hara K, Kashiwagi T, Hamada N, et al. Basic amino acids in the N-terminal half of the PB2 subunit of influenza virus RNA polymerase are involved in both transcription and replication. J Gen Virol. 2017;98(5):900–905. doi: 10.1099/jgv.0.000750
- Whelan M, Pelchat M. Role of RNA polymerase II promoter-proximal pausing in viral transcription. Viruses. 2022;14(9):2029. doi: 10.3390/v14092029
- Li B, Su G, Xiao C, et al. The PB2 co-adaptation of H10N8 avian influenza virus increases the pathogenicity to chickens and mice. Transbound Emerg Dis. 2022;69(4):1794–1803. doi: 10.1111/tbed.14157

