ABSTRACT
Pathogenic bacteria have evolved many strategies to evade surveillance and attack by complements. Streptococcus suis is an important zoonotic pathogen that infects humans and pigs. Hyaluronidase (HylA) has been reported to be a potential virulence factor of S. suis. However, in this study, it was discovered that the genomic region encoding HylA of the virulent S. suis strain SC19 and other ST1 strains was truncated into four fragments when aligned with a strain containing intact HylA and possessing hyaluronidase activity. As a result, SC19 had no hyaluronidase activity, but one truncated HylA fragment, designated as HylS,’ directly interacted with complement C3b, as confirmed by western ligand blotting, pull-down, and ELISA assays. The deposition of C3b and membrane attack complex (MAC) formation on the surface of a HylS’-deleted mutant (ΔhylS’) was significantly increased compared to wild-type SC19. In human sera and whole blood, ΔhylS’ survival was significantly reduced compared to that in SC19. The resistance of ΔhylS’ to macrophages and human polymorphonuclear neutrophil PMNs also decreased. In a mouse infection model, ΔhylS’ showed reduced lethality and lower bacterial load in the organs compared to that of SC19. We conclude that the truncated hyaluronidase HylS’ fragment contributes to complement evasion and the pathogenesis of S. suis.
Introduction
After entering the host, pathogens evade multiple attacks by the immune system to survive. As an ancient mechanism, the complement system is one of the first lines of defence against pathogens, enabling rapid identification and elimination of evaders [Citation1]. The complement system consists of approximately 50 proteins in the plasma or associated with cell membranes, forming a tightly coupled and highly collaborative network. The functions of complement include immune surveillance, detection and elimination of microbes, mediation of inflammatory responses, and participation in many physiological pathways [Citation2,Citation3]. It also acts as a bridge between innate and acquired immunities [Citation4]. There are three complement activation pathways: the classical, alternative, and lectin pathways. Activation of one of the three pathways leads to the formation of a membrane attack complex (MAC) consisting of C5b, C6, C7, C8, and C9 proteins on the membrane of the target cell [Citation5]. The complement component C3 is a central element of the three complement pathways [Citation6]. Upon encountering pathogens or their components, the abundant plasma protein C3 is cleaved into C3b and C3a by C3 convertase, which ultimately leads to MAC formation on the target surface [Citation7]. Immuno-adhesion and opsonophagocytosis also require fragment C3b [Citation8]. C3b and iC3b are recognized by complement receptor 1 (CD35) and complement receptors 3 and 4 (CD11b/18 and CD11c/18), respectively, on the surface of phagocytes to facilitate the phagocytosis of immune complexes or target cells [Citation3,Citation9].
As described above, C3 and its fragments play essential roles in immune surveillance and clearance of pathogens, but microbes have evolved many escape strategies [Citation2], For example, the Sbi protein of Staphylococcus aureus can form a ternary complex with Factor H and C3b, which is a potent inhibitor of the alternative pathway [Citation10]. In addition, SplB secreted by S. aureus can degrade α-chains of C3b, thereby overcoming complement attack [Citation11]. Similarly, LytA murein hydrolase expressed by Streptococcus pneumoniae can cleave C3b and iC3b deposited on the bacterial surface, resulting in reduced opsonic activity and phagocytosis [Citation12].
Streptococcus suis is a major pathogen with multiple serotypes [Citation13]. As an emerging zoonotic pathogen that infects both swine and humans, S. suis serotype 2 is one of the most virulent and prevalent serotypes [Citation14]. Bacterial meningitis and streptococcal toxic shock-like syndrome (STSLS) are the main symptoms of humans infected with S. suis, however, endocarditis, cellulitis, peritonitis, arthritis, pneumonia, and other symptoms have also been reported [Citation15,Citation16] and [Citation17]. After S. suis invasion, an early burst of host inflammatory cytokines can lead to STSLS, with S. suis using various strategies to survive in the blood, escape polymorphonuclear leukocyte-mediated phagocytosis, and penetrate the blood–brain barrier (BBB) to cause meningitis [Citation18]. The important virulence factor suilysin (SLY) enhances the survival of S. suis in serum [Citation19]. S. suis expresses many factors that enable the evasion of the host immune system. For example, in our previous study, S. suis SntA was found to interact with complement C1q, thereby inhibiting C3 deposition and MAC formation on S. suis [Citation20]. Bacterial adenosine synthase converts adenosine monophosphate into adenosine, which is used by S. suis to evade phagocytosis in blood [Citation21]. Additionally, S. suis 5’-nucleotidase affects neutrophil activity through adenosine synthesis [Citation22].
Hyaluronic acid (HA) is a major component of the extracellular matrix (ECM) of eukaryotic cells and plays a key role in regulating inflammation [Citation23]. Bacterial hyaluronidase degrades HA into unsaturated disaccharides by cleaving β-1–4 glycosidic bonds, and has been reported as a virulence factor in various bacterial species by reducing the viscosity of the ECM and breaking down connective tissue [Citation24,Citation25]. Hyaluronidase facilitates the absorption and spread of bacterial toxins, leading to further bacterial infection and colonization of bacteria [Citation26–28].
The hyaluronidase from Streptococcus agalactiae (also known as group B Streptococcus or GBS),
which can cause serious illness and/or death in newborns, the elderly, and the immunocompromised [Citation29–31], can degrade HA and enhance pathogenesis [Citation32], and is also considered a virulence factor in other gram-positive bacteria [Citation33]. Hyaluronidase also contributes to the intracellular survival of Streptococcus agalactiae in macrophages by inhibiting inflammatory cytokine expression [Citation34]. Hyaluronidase of S. suis serotype 2 (HylA) interacts with the murine angiogenin inhibitor 1 (AI1) [Citation35]. However, in the genome of S. suis serotype 2 strain SC19, we identified a region encoding truncated HylA, the product of which had no hyaluronidase activity. A fragment named HylS’ in this region was confirmed to be a secretory protein that interacts with complement C3b. C3b and MAC deposition on the surface of the HylS’-deleted mutant was increased compared with wild-type strain SC19. The hylS’ mutant showed reduced survival in human serum, whole blood, and anti-phagocytic activity. The hylS’ mutant was also significantly attenuated in mice. These findings indicated that HylS’ plays a role in S. suis complement evasion and virulence.
Materials and methods
Strains, plasmids, primers, and culture conditions
The bacterial strains, plasmids, and primers used in the present study are listed in Table S1. S. suis serotype 2 strain SC19 was originally isolated from sick pigs during an epidemic outbreak in the Sichuan province of China in 2005 [Citation15]. S. suis strains were grown in Tryptic Soy Broth (TSB; BD 211,825) or on Tryptic Soy Agar (TSA; BD 221,185) plates supplemented with 5% newborn bovine serum (Sijiqing 22,011–8612) at 37°C. Escherichia coli DH5ɑ and BL21 (DE3) strains were grown in Luria-Bertani (LB; Hopebiol, HB0128) broth or on LB agar plates at 37°C. The RAW264.7 cells were cultured in DMEM supplemented with 10% (v/v) foetal bovine serum (FBS), 100 µg/mL ampicillin, and 100 μg/mL streptomycin at 37°C with 5% (v/v) CO2.
Construction of the mutants and complementary strains
The S. suis mutant strain ΔhylS’ was constructed by knocking out the gene using homologous recombination [Citation36]. The 1000 bp region upstream of the hylS’ gene (GenBank accession number: OR478962), followed by the erm expression cassette and the 1000 bp region downstream of the hylS’ gene were cloned into the BamHI/EcoRI digested product of the suicide vector pSET4S. Afterward the recombinant plasmid was transformed into SC19 and the strains were selected on TSA plates with spectinomycin. The gene deletion mutant was identified from the transformants using the primers listed in Table S1 to amplify the intact coding sequence and the internal fragment of hylS’ (ΔhylS’). The absence of hylS’ expression and expression of the upstream and downstream genes of ΔhylS’ were further confirmed using cDNA of the mutant using the primers listed in Table S1.
To construct the complementary strain of ΔhylS’ (CΔhylS’), hylS’ was amplified by PCR using SC19 genomic DNA as a template. The PCR product was ligated into the vector pSET2 [Citation37] linearized with the restriction enzymes BamHI and EcoRI. Finally, the recombinant plasmid pSET2-hylS’ was electro-transformed into the mutant ΔhylS’, which was confirmed by amplification of the intact coding sequence of hylS’ using the primers listed in Table S1.
For the construction of the mutant containing intact hylA instead of the original truncated region (h1215-h1212) in SC19, the 1000 bp sequence upstream of the h1215 gene of SC19, followed by the hylA amplified from the genomic DNA of S. suis 0895 [Citation38] and a 1000 bp sequence downstream of the h1212 gene of SC19, were cloned into the BamHI/EcoRI digested product of the suicide vector pSET4S. The recombinant plasmid was then electrotransformed into SC19 cells and the transformants were selected on TSA plates with spectinomycin. The mutated SC19 containing intact hylA in place of h1215-h1212 (SC19hylA) was identified by PCR using primer hylA-F/R. Compared with hylA, h1215–1212 has a 21bp insertion at position 192 (). The primers hylA-F contained 13 bp of the 21bp sequence. Colonies with no PCR product using primers hylA-F/R were selected as SC19hylA., which was subsequently confirmed by PCR using cDNA as template using primers hylA-F/R.
Figure 1. Sequence analysis of the genomic region encoding HylA of S. suis and hyaluronidase activities of HylA and the products of truncated HylA. (a) Analysis of nucleic acid sequence of the genomic region encoding HylA of S. suis. Upper panel shows the sequence comparison of S. suis SC19 and another clinical isolate 0895. Letters with yellow shading highlight the differences in these two sequences. The region encoding HylA in the SC19 genome was split into four genes (h1215, h1214, hylS’ and h1212) as indicated in the sequences and in the gene location figure in the lower panel. (b) Hyaluronidase activities of different S. suis strains. Compared with the strain 0895 with intact hylA and strain SC19hylA with intact hylA instead of truncated hylA, no halo around the growth area of SC19 could be observed, indicating that SC19 had no hyaluronidase activity. (c) Hyaluronidase activities of the proteins of H1212, HylS,’ H1214, H1215 and HylA when expressed in E. coli BL21. The genes encoding H1212, HylS,’ H1214, and H1215 were amplified from SC19 and the gene encoding intact HylA was amplified from 0895. The genes were cloned into plasmid pET28a and transformed into BL21. The hyaluronidase activities of BL21 containing different recombinant proteins were analysed. There was no halo around the growth area of BL21 and BL21 containing H1212, HylS,’ H1214 or H1215, indicating none of these proteins had hyaluronidase activity. BL21 containing intact HylA was used as the positive control. All the experiments were performed in triplicate.
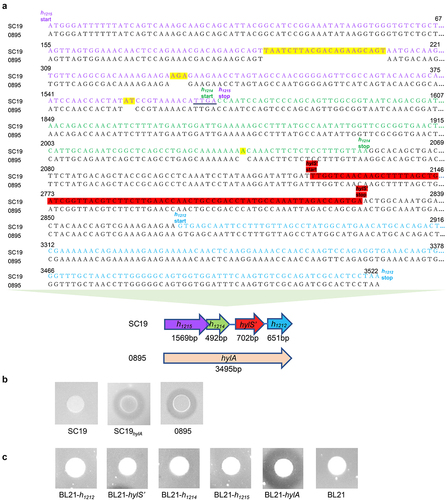
Plate assay for determination of hyaluronidase activity
Hyaluronidase activity was determined using a plate assay as described previously [Citation39]. Briefly, 10 µL of mid-log phase cultures of S. suis was spotted on Brain Heart Infusion (BHI; BD-Canada) agar plates supplemented with 0.04% (w/v) hyaluronic acid (Sigma-Aldrich 385,908) and 1% (w/v) bovine serum albumin (BSA). Plates were then incubated for 24 h at 37°C. Hyaluronidase activity was revealed by the addition of 2 M acetic acid for 3 min, which allowed precipitation of the BSA/hyaluronic acid complex, and the presence of a clear halo around bacterial growth indicated hyaluronidase activity.
To detect the hyaluronidase activities of E. coli expressing different recombinant proteins, the genes h1212, hylS’, h1214, and h1215 amplified from SC19 and hylA amplified from 0895 were cloned into pET28a and transformed into BL21. Hyaluronidase activity of the resultant strains was determined using the method described above.
Protein expression and purification
Recombinant H1212, HylS,’ H1214, H1215, HylA (S. suis strain 0895 [Citation38]; and C3b were expressed and purified. The full-length sequences of h1212 (GenBank accession number: OR478963), hylS’ (GenBank accession number: OR478962), h1214 (GenBank accession number: OR478961), and h1215 (GenBank accession number: OR478960) amplified from SC19 genomic DNA, and hylA (GenBank accession number: OR473667) amplified from strain 0895 were cloned into pGEX-6p. The sequence encoding the α-chain of C3b was amplified from human lung cDNA and cloned into the vector pET28a. Recombinant plasmids were transformed into BL21 (DE3) cells.
Protein expressions and purifications were performed as previously described [Citation40].
Recombinant proteins were expressed in E. coli BL21 (DE3) as GST-tagged proteins (GST – H1212, GST – HylS,’ GST – H1214, GST – H1215 and GST – HylA) or His-tagged C3b. Briefly, protein expression was induced with 0.5 mM isopropyl-β-D-thiogalactopyransoide (IPTG) at 16 °C for 20 h. The bacteria were centrifuged at 8000 g for 10 min, cells were harvested, and washed three times with PBS (2 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, and 3 mM KCl). Cells were resuspended in PBS and disrupted in an Ultrasonic Cell Crusher (Manufacturer, program). The protein fragments were separated by centrifugation. The washing buffer (50 mM Tris-HCl, 0.3 M NaCl, 10 mM EDTA, 10 mM DTT, and 0.5% Triton X-100) and resuspension buffer (50 mM Tris-HCl, 0.1 M NaCl, 10 mM EDTA, and 10 mM DTT) were used to resuspend the fragments. The precipitates were dissolved at a concentration of 30 mg/mL with the dissolution buffer (6 M gua-HCl, 10% glycerol, 50 mM Tris-HCl, 0.1 M NaCl, 10 mM EDTA, and 10 mM DTT). Next, the supernatants were obtained by centrifugation and mixed with the refolding buffer (0.1 M Tris-HCl, 0.4 M L-Arg HCl, 2 mM EDTA, 0.15% L-Glutathione reduced, and 0.03% L-Glutathione oxidized) for 8 h. The resulting protein liquids were concentrated and stored at − 80°C.
Preparation of anti-HylS’ antibody
Preparation of anti-HylS’ antibody was performed by three immunizations of recombinant HylS’ protein in Balb/c mice to obtain anti-serum from the mice blood [Citation41]. In brief, purified recombinant HylS’ protein was diluted to 300 μg/mL with PBS. Then, 2 mL of complete Freund’s adjuvant (Sigma-Aldrich, F5881) was mixed with an equal volume of diluted HylS’ protein. Subsequently, each mouse was injected intraperitoneally with the mixture containing 45 μg HylS’ protein. Two weeks later, a mixture of HylS’ added to incomplete Freund’s adjuvant (Sigma-Aldrich, F5506) was used for the second immunization of the mice. At week 5, each mouse was injected with 45 μg of protein with incomplete Freund’s adjuvant. At the sixth week, blood was collected and serum stored at −80°C.
Detection of subcellular localization of SC19 HylS’
The subcellular localization of HylS’ was determined as previously described [Citation42]. Briefly, bacterial cultures of SC19 or ΔhylS’ were collected by centrifugation, resuspended in 1 mL lysis buffer (0.05 M Tris – HCl, 2.5 mM EDTA, 0.1 M NaCl, 0.25% Triton X-100, pH 8.5 ~ 9.0) and boiled for 20 min to obtain cell lysates. The culture supernatants were filtered using filter membrane (Millipore, 0.22 µm, ISEQ00010) and concentrated to a final concentration of 10 mg/mL protein using ultrafiltration tubes, which were prepared as secretory proteins. Cell wall proteins were obtained from bacterial cultures by mixing with 1 mL of osmoprotective buffer (50 mM Tris – HCl, pH 7.3, 20% sucrose, 2.5 µM PMSF (phenyl methane sulphonyl fluoride)) containing mutanolysin (150 U/mL, Sigma-Aldrich, SRE0007) and incubated at 37°C for 2 h under continuous moderate agitation. Supernatants obtained after centrifugation at 12,000 g for 30 min. Cell lysate, secreted, and cell wall protein samples (50 µg of protein) were separated using 15% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (PVDF, Bio-Rad; 1620177). Mouse HylS’ polyclonal antibody was used to detect HylS,’ and HRP-conjugated goat anti-mouse IgG (Proteintech, SA00001–1) was used as the secondary antibody. Protein bands were visualized after adding a chemiluminescent substrate of HRP and detected using a ChemiDoc™ Touch Imaging System (Bio-Rad 1,708,370) in accordance with the manufacturer’s instructions. The cell wall anchored protein SntA of S. suis identified in our previous work was used as a control [Citation20].
Western ligand blotting
The western ligand blotting was performed as previously described [Citation43]. The proteins GST-H1212, HylS,’ GST-H1214, GST-H1215 and His-C3b were separated by denaturing 12% or 15% polyacrylamide gels (SDS-PAGE). GST and BSA were used as negative controls. Proteins were electrotransferred onto a PVDF, which was incubated in 5% skimmed milk for 2 h. After washing five times with TBST buffer (20 Mm Tris- HCl, 150 Mm NaCl, 0.05% Tween 20), His-C3b (10 µg/mL) was added to the membrane at 37 °C for 2 h, after which the membrane was washed a further five times. Mouse anti-human C3b monoclonal antibody (Proteintech 66,157–1-Ig) was used as the primary antibody, incubated at 37 °C for 2 h. After five TBST washes, HRP-conjugated goat-anti-Mouse IgG (Proteintech, SA00001–1) was added to the membrane and incubated at 37 °C for 2 h. Protein bands were visualized after adding a chemiluminescent substrate of HRP and detected using the ChemiDoc™ Touch Imaging System (Bio-Rad 1,708,370) in accordance with the manufacturer’s instructions.
Pull-down and western blot analysis
The pGEX-6p-hylS’ and pGEX-6p -hylA recombinant plasmids were transformed into E. coli BL21 (DE3) cells. The empty vector, pGEX-6p, served as the control. BL21 containing different expression vectors were grown in LB medium supplemented with 0.4 M glucose at 37°C to an OD600 of 0.5, and the temperature was raised to 42°C for 15 min. Afterward, 0.5 mM IPTG was added to induce the expression of the GST-tagged HylS’ or HylA at 16°C for 20 h. Bacteria were harvested by centrifugation, washed twice, and resuspended in PBS. The cells were disrupted with an Ultrasonic Homogenizer (Scientz, JY92-IIN), and the cell fragments were separated by centrifugation. The supernatants were added to Glutathione Sepharose® 4 Fast Flow resin (GE Healthcare 17,513,201), normal human serum from adult volunteers were added, then the resin was incubated at 4°C for 2 h. A PBS wash was carried out prior to the addition of elution buffer (150 mM NaCl, 50 mM pH 8.0 Tris-HCl, and 15 mM L-glutathione reduced). Samples were separated by 12% SDS-PAGE, proteins electro-transferred onto a PVDF membrane, incubated with 5% skimmed milk for 1 h, washed five times with TBST buffer, and incubated with mouse anti-human C3b monoclonal antibody (Proteintech 66,157–1-Ig) at 37 °C for 2 h. After washing another five times, HRP-conjugated goat-anti- mouse IgG (Proteintech, SA00001–1) was added and incubated at 37 °C for 2 h. Protein bands were visualized after addition of chemiluminescent substrate of HRP. A ChemiDoc™ Touch Imaging System (Bio-Rad 1,708,370) was used to detect the bands according to the manufacturer’s instructions.
C3b and HylS’ interaction detected by ELISA
The HylS’-binding assay was performed as follows. Different concentrations of His-C3b (0–7 µg/mL) were coated onto 96-well plates (BIOFIL, FEP-101-896) at 4°C for 12 h. Wells coated with BSA were used as a negative control. Wells were washed with PBST (PBS supplemented with 0.05% Tween-20), and 100 µL 1% BSA was added to each well for 2 h for blocking, followed by addition of 100 µL 5 µg/mL HylS’ to each well, and incubation at 37°C for 2 h. Mouse anti-HylS’ antibody was added to the wells and incubation continued at 37°C for 1 h. HRP-conjugated goat-anti-mouse IgG (Proteintech, SA00001–1) was used as the secondary antibody with incubation at 37°C for 1 h before TMB chromogenic solution (Beyotime, P0209) reaction for 30 min. Finally, the absorbance of the wells was measured at 630 nm. For the C3b binding assay, different concentrations of HylS’ or BSA (0–7 µg/mL) were used to coat 96-well plates. The same steps as the HylS’ binding assay were used, except that His-C3b was added instead of HylS’ and mouse anti-C3b antibody (Proteintech 66,157–1-Ig) was added instead of anti-HylS’ antibody.
Detection of C3b or MAC deposition on S. suis
C3b and MAC deposition on S. suis was detected as previously described [Citation44]. Briefly, bacteria were cultured in TSB overnight at 37°C and sub-cultured to mid-log phase. Bacterial cultures were harvested by centrifugation at 1556 g for 5 min, washed three times with PBS, and diluted to 5 × 107 CFU/mL with PBS. Supernatants containing proteins secreted from cognate bacterial cultures collected after centrifugation were filtered (Millipore, 0.22 µm, ISEQ00010) and concentrated to 10 mg/mL. Thereafter, 300 µL of bacterial suspensions, 300 µL fresh human serum, and 50 µL filtrates were mixed and incubated for 30 min at 37°C. Bacterial suspensions mixed with inactivated normal human serum served as negative controls. After incubation, the mixtures were centrifuged and washed thrice with PBS. To detect C3b, the mixtures were incubated with 300 µL of monoclonal anti-human C3b antibody (20 µg/mL, Abcam, ab17456) at room temperature for 30 min. Similarly, 300 µL of monoclonal anti-human-C5b-9 antibody (20 µg/mL; Abcam, ab66768) was used to detect MAC on the bacterial surface. FITC-conjugated goat-anti-mouse IgG (Proteintech, SA00003–1) was used as the secondary antibody and incubated at 37°C for 30 min. C3b or MAC deposition was detected using a FACSCalibur flow cytometer (BD Biosciences, USA). For each sample, 20,000 cells were randomly selected. A threshold value using the untreated bacteria was set as the negative control. The quantities of C3b deposition or MAC formation on bacteria are shown as geometric mean fluorescence intensities (GMF).
Bacterial survival in serum or whole blood
For the detection of bacterial survival in serum, strains grown in TSB at 37°C overnight were sub-cultured to mid-log phase. Cultures were diluted to 5 × 106 CFU/mL in PBS, and 20 µL of each diluted culture was mixed with 180 µL normal human serum and incubated at 37°C for 25 min. Inactivated serum was used as a negative control. Three replicates were performed for each strain. Live bacterial numbers were counted by plating serially diluted cultures on TSA plates. Survival percentages of bacteria were calculated as (CFUactive serum /CFUinactivated serum) × 100%. To detect of bacterial survival in whole blood, 50 µL of diluted mid-log phase bacterial culture (CFUinput) was incubated with 450 µL fresh human blood at 37°C for 60 min. Live bacterial numbers were counted by plating serially diluted cultures on TSA plates (CFUoutput). Survival percentages of bacteria were calculated as (CFUoutput/CFUinput) × 100%. The other steps were the same as those for the serum survival assay. The serum or whole blood used in this study were pooled samples obtained from multiple donors.
Phagocytosis assay
Macrophage phagocytosis assays were performed as previously described [Citation45]. RAW264.7 cells were incubated in 12-well plates until they were confluent single-layer, washed twice with PBS, and DMEM medium was added. Subsequently, log phase bacteria (CFUinput) were added to the plates (MOI = 10:1) and incubated for 60 min at 37°C. Three replicates were performed for each strain. The mixtures were washed twice with PBS and DMEM containing 100 µg/mL ampicillin was added for 1 h to eliminate extracellular bacteria. Saponin was added to lyse cells on ice. Live bacterial numbers were counted by plating diluted samples on TSA plates (CFUoutput). The ratio of engulfed bacteria was calculated as (CFUoutput/CFUinput) × 100%.
In addition, flow cytometry assays to quantify phagocytosis were also carried out as previously described [Citation46]. A GFP knock-in strain of SC19 was constructed. The ORF of GFP together with eno promoter sequences upstream of GFP were inserted into the intergenic region between the gene B9H01_RS02005 and B9H01_RS02010 in the genome of SC19 (SC19GFP). Then, ΔhylS’ and CΔhylS’ were also constructed using SC19GFP as parental strain. Log phase bacteria carrying GFP (CFUinput) were added to the plates containing RAW264.7 cells (MOI = 10:1) and incubated for 40 min at 37°C. GFP-negative bacteria (SC19) and RAW264.7 cells were used as negative controls. The wells were washed twice with PBS, then DMEM containing 100 µg/mL ampicillin was added for 1 h to eliminate the extracellular bacteria. Thereafter, the wells were washed twice with PBS, and the cells were harvested and fixed for 30 min with 4% paraformaldehyde at 4°C. Finally, the cells were applied to a FACSCalibur flow cytometer (BD Biosciences, USA). For each sample, 2,000 cells were randomly selected. A threshold value was set using the negative control, the cells without bacteria. The quantities of the cells containing bacteria expressing GFP are shown as geometric mean fluorescence intensities.
The survival of bacteria in the presence of PMNs was determined as previously described [Citation20]. A human peripheral blood PMN isolation kit (Solarbio, P9040) was used to extract neutrophils from human blood. Log-phase bacterial cultures were washed twice with PBS, and bacteria were diluted in RPMI-1640 medium (Hyclone, SH30809.01) to 5 × 107 CFU/ml. Next, 100 μL bacterial dilutions, 100 μL RPMI-1640 medium, 200 μL human normal serum, and 500 µL PMNs were mixed and incubated at 37°C in 5% CO2 for 30 min. Inactivated serum was used as a negative control. Three replicates were performed for each strain. Finally, 100 µL of saponin was added to each sample, which was then placed on ice for 10 min. The number of live bacteria was determined by plating the diluted cultures on TSA plates. The ratio of live bacteria was calculated as (CFUactive serum /CFUinactivated serum) × 100%.
Mouse infection experiments
To evaluate the lethality of different S. suis strains, 24 female 5-week-old SPF Kunming mice (eight mice per group) were intraperitoneally infected with 2 × 109 CFU/mouse of SC19, ΔhylS’ or physiological saline as a negative control. Mortality was recorded within 7 days after inoculation. To determine the bacterial loads in tissues infected with different S. suis strains, 48 female 5-week-old SPF Kunming mice were intraperitoneally infected with 5 × 108 CFU/mouse SC19 and ΔhylS’ (24 mice per group). At 6, 12, 24, and 48 h post-infection, six mice per group were sacrificed at each time point. Mouse organs were weighed, and bacterial numbers in the blood, lungs, spleen, and brain were counted by plating serially diluted homogenates onto TSA plates, and CFUs were determined after overnight incubation.
Statistical analysis
GraphPad Prism 8 software was used for statistical analysis. Unpaired one-tailed Student’s t-test was used to analyse the differences between the two groups from the results of protein interaction detected by ELISA, detection of deposition of C3b or MAC, survival in serum or whole blood, phagocytosis assay and bacterial loads in mouse tissues. The Log Rank test was used to analyse the survival rates between different groups in the mice infection assay. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, p > 0.05.
Results
HylS’ of S. suis SC19 had no hyaluronidase activity
Hyaluronidase has been reported as a virulence factor in various bacterial species because of its ability to degrade hyaluronic acid [Citation47,Citation48]. We aligned the nucleic acid sequences putatively encoding hyaluronidase from SC19 with those of other S. suis strains. The results showed that the region putatively encoding hyaluronidase from SC19 had four insertions compared to that from clinical isolate 0895, which had 21, 3, 2, and 1 bp insertions at positions 192, 330, 1554, and 2036, respectively (). The sequence of SC19 was identical to that of strains P1/7, P517/03P4, and MNCM01. Strain 0895 had the same nucleic acid sequence as strains MNCM04, MGGUS5, and MGGUS9. These results indicated that the region encoding the hyaluronidase-like fragment in SC19 was divided into four genes. Additionally, we confirmed that the four truncated genes were co-transcribed in an operon (Fig. S1). The relative positions of the four genes are shown in . These genes were named h1215 (1569 bp), h1214 (492 bp), hylS’ (702 bp), and h1212 (651 bp) ().
Then, the genes h1212, hylS’, h1214 and h1215 were cloned into expression vectors, enabling the expression and purification of the recombinant proteins H1212, HylS,’ H1214, and H1215, respectively. The hyaluronidase activities of SC19 and 0895 and the recombinant proteins H1212, HylS,’ H1214, and H1215 of S. suis were assessed. No visible halo was observed around the growth area of SC19 on the plate containing hyaluronic acid, indicating that SC19 had no hyaluronidase activity (). As controls, strain 0895 and a self-constructed mutated SC19 (SC19hylA, SC19 containing intact hylA in place of four truncated genes) displayed a clear halo, i.e., the dark grey circle around the bacteria (), indicating the hyaluronidase activity of intact HylA. There were no halos around E. coli expressing HylS,’ H1212, H1214 or H1215 on plates containing hyaluronic acid (). In contrast, E. coli expressing HylA at 0895 exhibited hyaluronidase activity (). Taken together, these results indicated that S. suis SC19 and its proteins H1212, HylS,’ H1214 and H1215 had no hyaluronidase activity.
HylS’ of S. suis SC19 is a secretory protein interacting with complement C3b
Western ligand blotting and pull-down assays were used to determine whether HylS’ or its adjacent associated fragments H1212 and H1214 and H1215 or HylA from S. suis 0895 interacted with human complement C3b. The results revealed an interaction between HylS,’ HylA, and C3b (). Fragments H1212, H1214 and H1215 did not bind to C3b. The controls, GST, the expression tags of HylS’ and HylA, and BSA had no interaction with C3b (). Additionally, the results of the pull-down assays were consistent with those of the western ligand blotting. After SDS-PAGE, HylS’ and HylA were associated with a band corresponding to the size of C3b (). This band was confirmed to be C3b by western blotting (). Thereafter, ELISA was used to further confirm the interaction of C3b with HylS,’ which showed that C3b could bind to HylS’ in a dose-dependent manner in HylS’-coated plates (). Similarly, C3b could bind to HylS’ in a dose-dependent manner in C3b-coated plates (). The negative control, BSA, did not show any binding ability to C3b or HylS’ (). Altogether, these results show that S. suis HylS’ interacts with human complement C3b.
Figure 2. HylS’ and HylA interact with complement C3b. (a) Western ligand blotting was used to detect interaction of H1212, HylS,’ H1214, H1215 or HylA with C3b. Purified recombinant proteins GST- H1212, GST-HylS,’ GST-H1214, GST-H1215, and GST-HylA were separated by SDS-PAGE. One gel was visualized by Coomassie blue staining (a). The proteins present in a separate identical gel were transferred onto a PVDF membrane, incubated with C3b, then detected with anti-C3 antibody followed by incubation with HRP-conjugated goat-anti-mouse IgG (b). BSA and GST were used as negative controls. The final lane was protein His-C3b. (b) Pull-down assay was used to detect the interaction of HylS’ or HylA with C3b. Samples from bacterial cells expressing different recombinant proteins (100 μg per sample) were loaded onto glutathione resin columns, incubated with normal human serum (NHS), and analysed by SDS-PAGE after Coomassie blue staining (a) and western blotting using anti-C3b antibody (b). Protein His-C3b was used as the control. (c) ELISA detection of interaction of HylS’ and C3b. C3b was applied to HylS’- or BSA-coated plates to determine the C3b binding capacity (a). HylS’ was applied to C3b- or BSA-coated plates to determine the HylS’ binding capacity (b). Unpaired one-tailed Student’s t-test was used to do statistical analysis, ****p < 0.0001.
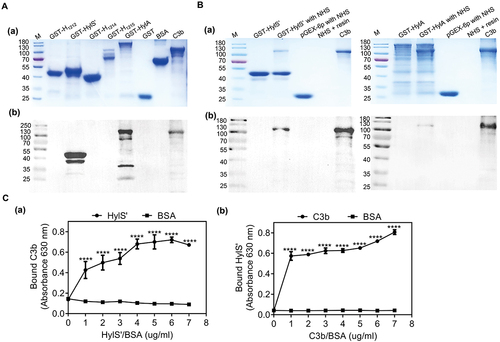
To determine the subcellular localization of S. suis HylS’ (27kDa), the HylS’ polyclonal antibody reactivity of different bacterial cellular components was assessed by western blotting. Anti-HylS’ reactivity was observed in concentrated culture supernatants, but not in cell lysates or cell wall proteins (), indicating that HylS’ is a secretory protein in S. suis. Compared with the parental strain SC19, the HylS’ band could not be detected in the culture supernatants of the gene-depleted mutant of hylS’ (ΔhylS’) (). The cell wall anchored protein SntA (91kDa) used as a control [Citation20] was observed in cell lysates and cell wall protein extracts, but not in culture supernatants ().
Figure 3. HylS’ is a secretory protein. The subcellular localization of HylS’ was determined by immunodetection of different components of bacteria. (a) Cell lysates, secreted proteins, and cell wall components were prepared from S. suis SC19 and subjected to western blot analysis using mouse anti-HylS’ polyclonal antibody, followed by HRP-conjugated goat-anti-mouse IgG. The band showed the size of protein HylS’ (27 kDa). (b) The secreted proteins of SC19 and the mutant ΔhylS’ were prepared and subjected to western blot analysis using mouse anti-HylS’ polyclonal antibody, followed by HRP-conjugated goat-anti-mouse IgG. The band showed the size of protein HylS’ (27 kDa). (c) The known cell wall anchored protein SntA was used as a control. Cell lysates, secreted proteins and cell wall components were prepared from S. suis SC19 and subjected to western blot analysis using mouse anti-SntA polyclonal antibody, followed by HRP-conjugated goat-anti-mouse IgG. The bands showed the size of SntA (91 kDa).
HylS’ inhibits C3b and MAC deposition on S. suis
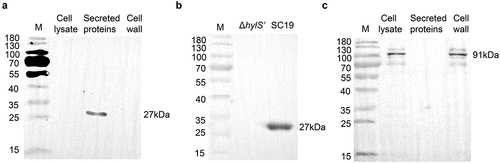
To further study the role of HylS’ in the immune evasion and virulence, SC19, ΔhylS’ and complementary strain (CΔhylS’) were used in the further studies (Fig. S2). SC19, ΔhylS’, CΔhylS’ and SC19hylA were incubated with fresh human serum, and the amount of C3b bound to the surface of the bacteria was detected using a fluorescence-labelled C3b antibody. The mean geometric fluorescence intensity (GMF) illustrated that the amount of C3b conjugated with ΔhylS’ was higher than that of SC19 and CΔhylS’ (p < 0.0001, ). The C3b deposition index of ΔhylS’ was significantly higher than those of SC19 and CΔhylS’ (). Furthermore, the formation of MAC on bacteria was measured using fluorescently labelled monoclonal anti-C5b-9 antibody. The GMF detected for ΔhylS’ was higher than that of SC19 and CΔhylS’ (p < 0.0001, ). The MAC deposition index of ΔhylS’ was also significantly higher than that of SC19 and CΔhylS’ (). In addition, increased deposition of C3b and MAC was found on the surface of SC19hylA compared with SC19, but was much lower than that of ΔhylS’ (p < 0.001, ). Taken together, these results show that the presence of HylS’ inhibits the deposition of C3b and MAC on S. suis.
Figure 4. HylS’ inhibits C3b deposition and MAC on S. suis. (a) Flow cytometry histogram showing C3b deposition on S. suis. The same amount of SC19, SC19hylA, ΔhylS’, and CΔhylS’ were incubated with normal human serum and bacterial culture supernatants of each strain. The bacteria incubated with C3b antibody were mixed with FITC-conjugated goat-anti-mouse IgG, then washed and detected by flow cytometry analysis. (b) Histogram showing MAC formation on S. suis. The same amount of SC19, SC19hylA, ΔhylS’, and CΔhylS’ were incubated with normal human serum and bacterial culture supernatants of each strain were subjected to flow cytometry analysis. C5b-9 antibody was used as the first antibody and FITC-conjugate goat-anti-mouse IgG as the secondary antibody. (c-d) Geometric mean fluorescence intensity (GMF) values of C3b deposition (c) and MAC formation (d) on S. suis. Data are shown as mean values ± SD from three independent experiments. Statistical analyses were performed using the unpaired one-tailed Student’s t-test, ****p < 0.0001; ***p < 0.001.
HylS’ enhances the survival ability of S. suis in serum and blood, and resistance to phagocytosis
Deposition of C3b on the surface of bacteria can affect bacterial survival in serum and blood. Thus, SC19, ΔhylS’ and CΔhylS’ were incubated with fresh human serum or blood to evaluate whether HylS’ affects the survival of S. suis in the serum and/or blood. The survival of SC19 in human serum was determined as 95 ± 7% after 25 min incubation. In comparison, the survivability of ΔhylS’ cells decreased to 74 ± 1% (). In addition, the survival of SC19 in human whole blood was 105 ± 9% after 30 min, but the survival percentage of ΔhylS’ was only 62 ± 9% (). There was no significant difference between SC19 and CΔhylS’ in the serum or blood (). Hence, the presence of HylS’ significantly improved the ability of S. suis SC19 to survive in human serum and blood samples.
Figure 5. HylS’ enhances the survivability of S. suis in serum, blood and resistance to phagocytosis. (a) Survival of SC19, ΔhylS’ and CΔhylS’ in human serum. The same amount of bacteria were incubated with human serum for 25 min, and the bacterial numbers were determined by viable counting after serial dilution and plating on TSA plates. Inactivated serum was used as a negative control. Survival percentages of bacteria were calculated as (CFUactive serum /CFUinactivated serum) × 100%. (b) Survival of SC19, ΔhylS’ and CΔhylS’ in whole blood. The same amount of bacteria (CFUinput) were incubated with heparinized human whole blood for 60 min, and the bacterial numbers were determined (CFUoutput). Survival percentages of bacteria were calculated as (CFUoutput/CFUinput) × 100%. (c) The ability of SC19, ΔhylS’ and CΔhylS’ to resist phagocytosis. The same amount of bacteria (CFUinput) were incubated with mouse macrophage RAW264.7 cells at a MOI of 1:10 (cell to bacteria). RAW264.7 cells were lysed with saponin and the bacterial numbers were determined (CFUoutput). The relative number of live bacteria was calculated as (CFUoutput/CFUinput) × 100%. (d) The ability of SC19, ΔhylS’ and CΔhylS’ to resist neutrophils-mediated killing. The same amount of bacteria were incubated with human neutrophils (PMNs) separated from fresh human blood at a MOI of 1:10 (cells to bacteria) together with human normal serum or heat-inactivated serum. PMNs were lysed and the bacterial numbers were determined. The relative number of live bacteria was calculated as (CFUserum/CFUinactivated serum) × 100%. Data are shown as mean values ± SD from three independent experiments. Statistical analyses were performed using the unpaired one-tailed Student’s t-test, ****p < 0.0001; ***p < 0.001; **p < 0.01; ns, p > 0.05.
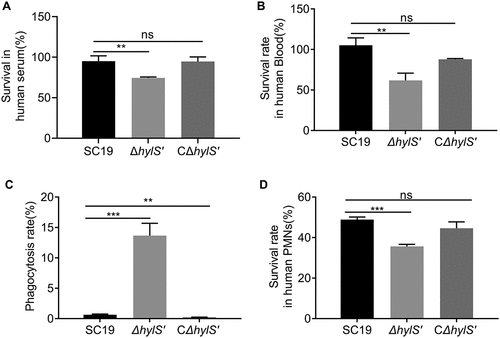
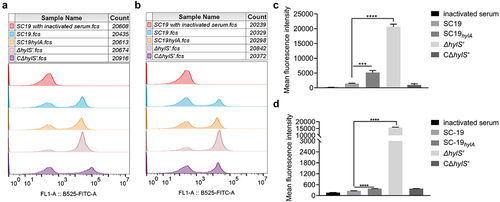
C3b can not only be deposited on the surface of bacteria to contribute to MAC activity but is also known to facilitate phagocytosis of pathogens. Thus, the ability of SC19, ΔhylS’ and CΔhylS’ strains to resist phagocytosis by macrophages and neutrophils was assessed. After incubation with macrophages, the phagocytosis percentage on ΔhylS’ was 13 ± 2%, which was significantly higher than that of SC19 and CΔhylS’ (p < 0.05) (). At the same time, by using the GFP expressing strains of SC19, ΔhylS’ and CΔhylS, the fluorescent bacteria inside the RAW264.7 cells were measured using flow cytometer. The mean geometric fluorescence intensities illustrated that the amount of ΔhylS’-GFP engulfed by RAW264.7 cells was significantly higher than that of SC19-GFP and CΔhylS’-GFP (p < 0.001, Fig. S3). Similarly, the survival percentage of ΔhylS’ was 35 ± 1% in neutrophils, which was significantly lower than that of SC19 and CΔhylS’ (p < 0.05) (). Therefore, the ability of S. suis SC19 to resist phagocytosis by macrophages and neutrophils was HylS’-dependent.
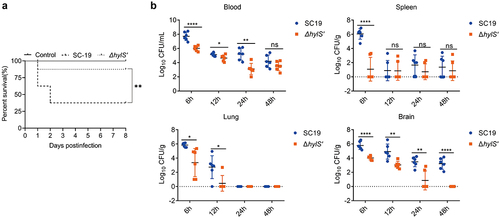
HylS’ contributes to the pathogenicity of S. suis in vivo
The survival of mice infected with equivalent doses of SC19 and ΔhylS’ was determined over 8 days. After two days post-infection, 37.5% of the mice infected with SC19 survived. In contrast, 87.5% of the mice in the ΔhylS’-infected group survived (p < 0.05) (). None of the mice in the negative control group died. Over the next 6 days, no mice in the infected and control groups died. Additionally, the bacterial loads in the tissues of the SC19 and ΔhylS’-infected groups were compared after a lower infection dose. The blood, lungs, spleens, and brains of the mice were collected at 6, 12, 24, and 48 h post-infection (hpi). At 6 hpi, the CFUs of SC19 in the blood, spleens, brains, and lungs were significantly higher than those of ΔhylS’ (p < 0.05). In the brains, from 6 to 48 hpi, the bacterial loads of ΔhylS’ mice were lower that of SC19 infected mice (p < 0.05) (). Taken together, these results indicated that HylS’ contributes to the pathogenicity of S. suis SC19 in mice.
Figure 6. HylS’ contributes to the pathogenicity of S. suis. (a) Survival curves of mice infected with SC19 and ΔhylS’. Two groups of mice (n = 8) were injected intraperitoneally with 2 × 109 CFU of SC19 or ΔhylS’. The Log Rank test was used to analyse the survival rates between different groups. **p < 0.01 (b) Bacterial loads in tissues of mice infected with SC19 and ΔhylS’. Two groups of mice (n = 6) were injected intraperitoneally with 5 × 108 CFU of SC19 or ΔhylS’. Data are shown as mean values ± SD. Statistical analyses were performed using the unpaired one-tailed Student’s t-test, ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, p > 0.05
Discussion
It is critical for bacteria to develop mechanisms to evade the host immune system to colonize and proliferate in the target host. The complement system is one such mechanism that enables the protection of humans and animals against invading pathogens. However, many pathogens have evolved strategies that interfere with their complement activity. Such complement evasion strategies include the production of proteases that degrade complement components, direct interactions with the complements, or the expression of complement modulators [Citation49]. The deployment of complement evasion strategies facilitates immune escape, enabling bacteria to cross the host mucosal epithelial barrier, enter the bloodstream, and spread into target organs such as the brain to cause disease [Citation50–52].
S. suis is an important zoonotic pathogen that has the ability to evade the host immune system. In the tonsils, S. suis survives by escaping phagocytosis by macrophages. In addition, the bacterium can invade the spleen, liver, kidney, and blood–brain barrier. Thus S. suis can cause meningitis, sepsis and STSLS [Citation15,Citation53]. Previous studies have reported the ability of S. suis to modulate and/or inhibit complement activity through the expression of proteins that directly interact with the complement components or receptors. These include the factor H-binding protein Fhb [Citation54], factor H-binding protein FHBP [Citation55], C1q binding protein SntA [Citation20], and cholesterol-binding cytolysin suilysin, which binds C3aR and C5aR [Citation56]. S. suis endopeptidase O (SsPepO) interacts with plasminogen, which is activated to produce plasmin to cleave C3b [Citation57], and immunoglobulin M-degrading enzyme (IdeSsuis) specifically cleaves porcine IgM to escape opsonization and complement-mediated killing [Citation58].
In this study, we identified a novel C3b interacting protein expressed by S. suis named HylS,’ which originates from hyaluronidase. Hyaluronidase is an important virulence factor in many pathogens. For example, hyaluronidase (HylB) in Group B Streptococcus enhances the virulence of this pathogen [Citation59]. It has been reported that co-administration of hyaluronidase with S. pneumoniae increases the number of mice who get meningitis [Citation60]. However, in this study, we found that S. suis strain SC19 had no hyaluronidase activity. Analysis of the DNA sequence in the putative hyaluronidase-encoding region in SC19 revealed four insertion mutations when compared to the equivalent intact hyaluronidase-encoding locus present in strain 0895, which possessed hyaluronidase activity (). The same mutations found in SC19 have also been described in S. suis strains belonging to the ST1 group, which are considered highly virulent and do not have hyaluronidase activity [Citation61]. Exogenous hyaluronic acid may enhance the virulence of S. suis strains without hyaluronidase activity by upregulating virulence genes [Citation61]. In addition, the absence of hyaluronidase activity in the majority of S. suis strains, which can infect humans and swine, indicates that hyaluronidase should not be considered as an essential virulence factor [Citation61,Citation62]. In the SC19 genome, the region encoding hyaluronidase was split into four genes, in which hylS’ was the third gene, 702 bp in length. We found that neither the entire region nor the four split genes showed hyaluronidase activity (). However, among the four genes, the expression product of hylS’ could interact with the host complement C3b (). The intact hyaluronidase from strain 0895 also bound to C3b, most likely because of the HylS’ component of the protein (). Since intact hyaluronidase HylA had both hyaluronidase activity and complement binding ability, it was difficult to distinguish between the two roles experimentally. The fragment HylS’ without hyaluronidase activity was used to evaluate its roles on complement evasion and pathogenicity. In a previous study, one fragment of HylA was described as interacting with a eukaryotic protein, angiogenin inhibitor 1, identified from a murine brain cDNA library [Citation35]. By comparing the sequences of primers used in that study, we found that the gene identified was most likely h1215, the first gene of the four truncated fragments of hylA. Together with the findings of our study, it can be speculated that the HylS’ fragment has other functions as well.
C3b is a key component of the complement system. Proteins synthesized by pathogens mainly inhibit the action of C3 in three ways: inhibition of C3 convertase formation, cleavage of C3b, or interaction with C3b to impact its function [Citation51]. For example, the extracellular fibrinogen-binding protein secreted by S. aureus interacts with C3b and directly impairs C3 convertase [Citation63]. The C3b-binding protein Ecb in S. aureus also blocks C3 convertases [Citation64]. The sialylated type III capsular polysaccharide of Group B Streptococcus prevents C3 deposition on the bacterial surface and phagocytic elimination [Citation65]. In this study, HylS’ was found to bind C3b as a secretory protein (). C3b deposition on the surface of the hylS’ deletion mutant was significantly increased compared to the wild-type strain (), which should be due to the lack of HylS’ protein in the secreted proteins of ΔhylS’ (). C3b-deposited bacteria are more susceptible to death. It has been recognized that the critical effect of complement to fight gram-positive bacteria is opsonization, by which C3 and C3b are deposited on bacteria to facilitate phagocytosis by PMNs, monocytes, and macrophages [Citation66]. In this study, ΔhylS’ was significantly more susceptible to phagocytosis by macrophages and PMNs ( and Fig S3AB). Complement receptor CR1 on the surfaces of macrophages and neutrophils engages with C3b resulting in opsonization [Citation67,Citation68]. Therefore, it is reasonable to infer that HylS’ protects S. suis from phagocytosis by interacting with C3b, probably by reducing the binding of C3b to receptors on phagocytes (). RAW macrophage phagocytosis experiments were carried out without any source of complement in the assay ( and Fig S3AB), suggesting that HylS’ may also participate in the evasion of phagocytosis via a complement-independent manner. Compared to the wild-type strain, there was a significant increase in MAC formation with ΔhylS’ (). MAC formation is the final step of the complement activation pathway, and results in bacterial lysis [Citation69]. The interaction of HylS’ with C3b may block the formation of C5 convertase, thereby affecting the deposition of C5b on the bacterial cell surface, resulting in a reduction in MAC formation, making SC19 less susceptible to complement attack (). Finally, it was discovered that the deletion of HylS’ reduced the survival of S. suis SC19 in human serum and blood (), lethality in mice, and bacterial loads in their organs (). Because HylS’ lacks hyaluronidase activity, its contribution to the full virulence of S. suis is most likely due to its ability to interact with C3b. Taken together, these results suggest that S. suis HylS’ interacts with complement C3b, possibly consuming C3b in the serum, leading to a series of biological effects, including reduced C3b deposition and phagocytosis of bacteria, and decreased MAC formation on bacteria, which could promote the survival of S. suis in the blood and enhance its virulence in vivo (). Intact HylA was also found to have C3b binding ability (). However, compared with the wild-type SC19 carrying HylS,’ the mutated SC19 carrying the intact HylA had increased deposition of C3b and MAC formation in S. suis (). We speculate that some lineages of S. suis have lost the ability to express hyaluronidase in the evolutionary process, and the tradeoff is that its mutated product HylS’ has a greater ability to bind C3b ().
Figure 7. Model of HylS’-mediated complement evasion of S. suis. In the genome of S. suis strain SC19, the hylS’ gene is derived from the hyaluronidase-encoding hylA gene: HylS’ does not have hyaluronidase activity. HylS’ can interact with host complement C3b, leading to the inhibition of C3b deposition on the bacterial surface, blockade of C5 convertase synthesis, and ultimately less MAC formation on the bacterial surface. Bacteria with reduced C3b deposition are less likely to be recognized by receptors on the surface of macrophages and neutrophils, and are more resistant to phagocytosis. This strategy of complement evasion promotes bacteria to further invade host tissues and organs.
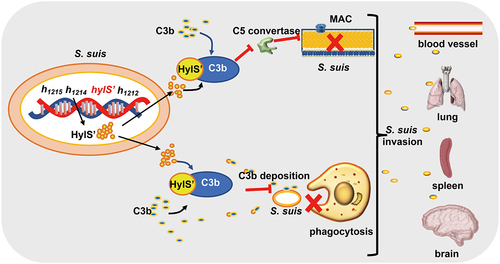
In conclusion, this study demonstrated that the genomic region encoding hyaluronidase in S. suis SC19 was truncated into four fragments and that the strain had lost hyaluronidase activity. The product of one fragment, HylS,’ binds to host C3b, leading to reduced deposition of C3b and MAC on S. suis. The contribution of HylS’ to SC19 survival in serum and blood, and resistance to PMNs was consistent with the block of deposition of C3b and MAC on bacteria. Virulence of the HylS’-deleted mutant was also significantly reduced. Thus, we discovered a novel complement evasion protein of S. suis and identified possibly underestimated functions of truncated fragments of hyaluronidase.
Supplemental Material
Download MS Excel (15.8 KB)Figure S1.jpg
Download JPEG Image (2.4 MB)Figure S2.jpg
Download JPEG Image (12.6 MB)Figure S3.jpg
Download JPEG Image (4.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
The data that support the findings of this study are openly available in Figshare at https://figshare.com/s/9c2cd1f3825b63f48891.
Supplemental data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2024.2306691
Additional information
Funding
References
- Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J Immunol. 2013;190(8):3831–16. doi: 10.4049/jimmunol.1203487
- Ricklin D, Reis ES, Mastellos DC, et al. Complement component C3 – the “Swiss army knife” of innate immunity and host defense. Immunol Rev. 2016;274(1):33–58. doi: 10.1111/imr.12500
- Reichhardt MP, Merib S. Intracellular complement activation—An alarm raising mechanism? Semin Immunol. 2018;38:54–62. doi: 10.1016/j.smim.2018.03.003
- Rambach G, Wurzner R, Speth C. Complement: an efficient sword of innate immunity. Contrib Microbiol. 2008;15:78–100. doi: 10.1159/000136316
- Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011;343(1):227–235. doi: 10.1007/s00441-010-1034-0
- Ricklin D, Hajishengallis G, Yang K, et al. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923
- Gadjeva M. The complement system. Overview. Methods Mol Biol. 2014;1100:1–9. doi: 10.1007/978-1-62703-724-2_1
- Sharma S, Bhatnagar R, Gaur D. Complement evasion strategies of human pathogenic bacteria. Indian J Microbiol. 2020;60(3):283–296. doi: 10.1007/s12088-020-00872-9
- Bajic G, Yatime L, Sim RB, et al. Structural insight on the recognition of surface bound opsonin by integrin I domain of complement receptor 3. Proc Natl Acad Sci, USA. 2013;110(41):16426–16431. doi: 10.1073/pnas.1311261110
- Haupt K, Reuter M, van den Elsen J, et al. The Staphylococcus aureus protein sbi acts as a complement inhibitor and forms a tripartite complex with host complement factor H and C3b. PLoS Pathog. 2008;4(12):e1000250. doi: 10.1371/journal.ppat.1000250
- Dasari P, Nordengrun M, Vilhena C, et al. The protease SplB of Staphylococcus aureus targets host complement components and inhibits complement-mediated bacterial opsonophagocytosis. J Bacteriol. 2021;204(1):e00184–21. JB0018421. doi: 10.1128/JB.00184-21
- Ramos-Sevillano E, Urzainqui A, Campuzano S, et al. Pleiotropic effects of cell wall amidase LytA on Streptococcus pneumoniae sensitivity to the host immune response. Infect Immun. 2015;83(2):591–603. doi: 10.1128/IAI.02811-14
- Zheng H, Qiu X, Roy D, et al. Genotyping and investigating capsular polysaccharide synthesis gene loci of non-serotypeable Streptococcus suis isolated from diseased pigs in Canada. Vet Res. 2017;48(1):10. do i:
- Gottschalk M, Xu J, Calzas C, et al. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010;5(3):371–391. doi: 10.2217/fmb.10.2
- Yu H, Jing H, Chen Z, et al. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12(6):914–920. doi: 10.3201/1206.051194
- Mai NT, Hoa NT, Nga TV, et al. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. 2008;46(5):659–667. doi: 10.1128/.00417-22
- Staats JJ, Feder I, Okwumabua O, et al. Streptococcus suis: past and present. Vet Res Commun. 1997;21(6):381–407. doi: 10.1023/A:1005870317757
- Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol. 2000;76:259–272. doi: 10.1016/S0378-1135(00)00250-9
- Lecours MP, Gottschalk M, Houde M, et al. Critical role for streptococcus suis cell wall modifications and suilysin in resistance to complement-dependent killing by dendritic cells. J Infect Dis. 2011;204(6):919–929. doi: 10.1093/infdis/jir415
- Deng SM, Xu T, Fang Q, et al. The surface-exposed protein SntA contributes to complement evasion in zoonotic Streptococcus suis. Front Immunol. 2018;9:1063–1063. doi: 10.3389/fimmu.2018.01063
- Liu P, Pian Y, Li X, et al. Streptococcus suis adenosine synthase functions as an effector in evasion of PMN-mediated innate immunity. J Infect Dis. 2014;210(1):35–45. doi: 10.1128/.00417-22
- Dai J, Lai L, Tang H, et al. Streptococcus suis synthesizes deoxyadenosine and adenosine by 5’-nucleotidase to dampen host immune responses. Virulence. 2018;9(1):1509–1520. doi: 10.1080/21505594.2018.1520544
- Litwiniuk M, Krejner A, Speyrer MS, et al. Hyaluronic acid in inflammation and tissue regeneration. Wounds Compend Clin Res Pract. 2016;28:78–88.
- Stern R, Jedrzejas MJ. Hyaluronidases: their genomics, structures, and mechanisms of action. Chem Rev. 2006;106(3):818–839. doi: 10.1021/cr050247k
- Tao L, Song F, Xu N, et al. New insights into the action of bacterial chondroitinase AC I and hyaluronidase on hyaluronic acid. Carbohydr Polym. 2017;158:85–92. doi: 10.1016/j.carbpol.2016.12.010
- Li S, Jedrzejas MJ. Hyaluronan binding and degradation by Streptococcus agalactiae hyaluronate lyase. J Biol Chem. 2001;276(44):41407–41416. doi: 10.1074/jbc.M106634200
- Marion C, Stewart JM, Tazi MF, et al. Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect Immun. 2012;80(4):1390–1398. doi: 10.1128/IAI.05756-11
- Ibberson CB, Jones CL, Singh S, et al. Staphylococcus aureus hyaluronidase is a CodY-regulated virulence factor. Infect Immun. 2014;82(10):4253–4264. doi: 10.1128/IAI.01710-14
- Mullaney DM. Group B streptococcal infections in newborns. J ObstetGynecol Neonatal Nurs. 2001;30(6):649–658. doi: 10.1111/j.1552-6909.2001.tb00012.x
- Kothari NJ, Morin CA, Glennen A, et al. Invasive group B Streptococcal disease in the elderly, Minnesota, USA, 2003-2007. Emerg Infect Dis. 2009;15(8):1279–1281. doi: 10.3201/eid1508.081381
- Francois Watkins LK, McGee L, Schrag SJ, et al. Epidemiology of invasive group B streptococcal infections among nonpregnant adults in the United States, 2008-2016. JAMA Intern Med. 2019;179(4):479–488. doi: 10.1128/spectrum.00417-22
- Kolar SL, Kyme P, Tseng CW, et al. Group B Streptococcus evades host immunity by degrading hyaluronan. Cell Host Microbe. 2015;18(6):694–704. doi: 10.1128/spectrum.00417-22
- Hynes WL, Walton SL. Hyaluronidases of gram-positive bacteria. FEMS Microbiol Lett. 2000;183(2):201–207. doi: 10.1111/j.1574-6968.2000.tb08958.x
- Wang ZF, Guo CM, Xu YN, et al. Two novel functions of hyaluronidase from streptococcus agalactiae are enhanced intracellular survival and inhibition of proinflammatory cytokine expression. Infect Immun. 2014;82(6):2615–2625.
- Wu T, Yuan FY, Chang HT, et al. Identification of a novel angiogenin inhibitor 1 and its association with hyaluronidase of Streptococcus suis serotype 2. Microbial Pathogenesis. 2010;49(1–2):32–37. doi: 10.1016/j.micpath.2010.03.002
- Takamatsu D, Osaki M, Sekizaki T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid. 2001;46(2):140–148. doi: 10.1006/plas.2001.1532
- Takamatsu D, Osaki M, Sekizaki T. Construction and characterization of streptococcus suis–Escherichia coli shuttle cloning vectors. Plasmid. 2001;45(2):101–113. doi: 10.1006/plas.2000.1510
- Zou G, Zhou J, Xiao R, et al. Effects of environmental and management-associated factors on prevalence and diversity of Streptococcus suis in clinically healthy pig herds in China and the United Kingdom. Appl Environ Microbiol. 2018;84(8):e02590–e02517. doi: 10.1128/AEM.02590-17
- Smith RF, Willett NP. Rapid plate method for screening hyaluronidase and chondroitin sulfatase-producing microorganisms. Appl Environ Microbiol. 1968;16(9):1434–1436. doi: 10.1128/am.16.9.1434-1436.1968
- Palmer I, Wingfield PT. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Curr Protoc Protein Sci. 2012;70(1):6–3. doi: 10.1002/0471140864.ps0603s70
- Hardy CM, Mobbs KJ. Expression of recombinant mouse sperm protein sp56 and assessment of its potential for use as an antigen in an immunocontraceptive vaccine. Mol Reprod Dev. 1999;52:216–224. doi: 10.1002/(SICI)1098-2795(199902)52:2<216:AID-MRD13>3.0.CO;2-R
- Stalhammar-Carlemalm M, Stenberg L, Lindahl G. Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J Exp Med. 1993;177(6):1593–1603. doi: 10.1084/jem.177.6.1593
- McDevitt D, Francois P, Vaudaux P, et al. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x
- Agarwal V, Sroka M, Fulde M, et al. Binding of Streptococcus pneumoniae endopeptidase O (PepO) to complement component C1q modulates the complement attack and promotes host cell adherence. J Biol Chem. 2014;289(22):15833–15844. do i:
- Carlsson F, Berggard K, Stalhammarcarlemalm M, et al. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med. 2003;198(7):1057–1068. doi: 10.1084/jem.20030543
- Boero E, Brinkman I, Juliet T, et al. Use of flow cytometry to evaluate phagocytosis of Staphylococcus aureus by human neutrophils. Front Immunol. 2021;12:635825. doi: 10.3389/fimmu.2021.635825
- Makris G, Wright JD, Ingham E, et al. The hyaluronate lyase of Staphylococcus aureus – a virulence factor? Microbiology. 2004;150(6):2005–2013. doi: 10.1099/mic.0.26942-0
- Hart ME, Hart MJ, Roop AJ. Genotypic and phenotypic assessment of hyaluronidase among type strains of a select group of staphylococcal species. Int J Microbiol. 2009;2009:614371. doi: 10.1155/2009/614371
- Yu YF, Wang J, Han R, et al. Mycoplasma hyopneumoniae evades complement activation by binding to factor H via elongation factor thermo unstable (EF-Tu). Virulence. 2020;11(1):1059–1074. do i:
- Rooijakkers SH, van Strijp JA. Bacterial Complement Evasion. Mol Immunol. 2007;44(1–3):23–32. doi: 10.1016/j.molimm.2006.06.011
- Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6(2):132–42. doi: 10.1038/nrmicro1824
- Blom AM, Hallstrom T, Riesbeck K. Complement evasion strategies of pathogens—acquisition of inhibitors and beyond. Mol Immunol. 2009;46(14):2808–2817. doi: 10.1016/j.molimm.2009.04.025
- Baums CG, Kock K, Beineke A, et al. Streptococcus suis Bacterin and subunit vaccine immunogenicities and protective efficacies against serotypes 2 and 9. Clin Vaccine Immunol. 2009;16(2):200–208. doi: 10.1128/CVI.00371-08
- Li XQ, Liu P, Gan SZ, et al. Mechanisms of host-pathogen protein complex formation and bacterial immune evasion of Streptococcus suis protein Fhb. J Biol Chem. 2016;291(33):17122–17132. doi: 10.1074/jbc.M116.719443
- Li Q, Ma C, Fu Y, et al. Factor H specifically capture novel factor H-binding proteins of Streptococcus suis and contribute to the virulence of the bacteria. Microbiol Res. 2017 [cited 2016 Nov 24];196:17–25. doi: 10.1016/j.micres.2016.11.011
- Deng SM, Zhao LY, Zhu JQ, et al. Complement C3aR/C5aR-binding protein Suilysin of Streptococcus suis contributes to monocyte chemotaxis. Vet Microbiol. 2020;242:108599. doi: 10.1016/j.vetmic.2020.108599
- Zhou Y, Yan K, Sun CF, et al. Binding of Plasminogen to Streptococcus suis Protein Endopeptidase O Facilitates Evasion of Innate Immunity in Streptococcus suis. Front Microbiol. 2021;12:694103. doi: 10.3389/fmicb.2021.694103
- Rungelrath V, Weisse C, Schutze N, et al. IgM cleavage by Streptococcus suis reduces IgM bound to the bacterial surface and is a novel complement evasion mechanism. Virulence. 2018;9(1):1314–1337. doi: 10.1080/21505594.2018.1496778.
- Gendrin C, Vornhagen J, Armistead B, et al. A nonhemolytic group B Streptococcus strain exhibits hypervirulence. J Infect Dis. 2018;217:983–987. doi: 10.1093/infdis/jix646
- Zwijnenburg PJ, van der Poll T, Florquin S, et al. Experimental pneumococcal meningitis in mice: a model of intranasal infection. J Infect Dis. 2001;183:1143–1146. doi: 10.1086/319271
- Haas B, Vaillancourt K, Bonifait L, et al. Hyaluronate lyase activity of Streptococcus suis serotype 2 and modulatory effects of hyaluronic acid on the bacterium’s virulence properties. BMC Res Notes. 2015;8(1):722–722. doi: 10.1186/s13104-015-1692-9
- King SJ, Allen AG, Maskell DJ, et al. Distribution, genetic diversity, and variable expression of the gene encoding hyaluronate lyase within the Streptococcus suis population. J Bacteriol. 2004;186(14):4740–4747. doi: 10.1128/JB.186.14.4740-4747.2004
- Koch TK, Reuter M, Barthel D, et al. Staphylococcus aureus proteins Sbi and Efb recruit human plasmin to degrade complement C3 and C3b. PLoS One. 2012;7(10):e47638. doi: 10.1371/journal.pone.0047638
- Jongerius I, Garcia BL, Geisbrecht BV, et al. Convertase inhibitory properties of staphylococcal extracellular complement-binding protein. J Biol Chem. 2010;285(20):14973–14979. doi: 10.1074/jbc.M109.091975
- Marques MB, Kasper DL, Pangburn MK, et al. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B Streptococci. Infect Immun. 1992;60(10):3986–3993. doi: 10.1128/iai.60.10.3986-3993.1992
- Brown EJ. Interaction of gram-positive microorganisms with complement. Curr Top Microbiol Immunol. 1985;121:159–187. doi: 10.1007/978-3-642-45604-6_8
- Law SK. C3 receptors on macrophages. J Cell Sci Suppl. 1988(9Supplement_9):67–97. doi: 10.1242/jcs.1988.supplement_9.4
- O’Shea JJ, Brown EJ, Seligmann BE, et al. Evidence for distinct intracellular pools of receptors for C3b and C3bi in human neutrophils. J Immunol. 1985;134(4):2580–2587. doi: 10.1056/NEJM200104053441406
- Walport MJ, Mackay IR, Rosen FS. Complement. first of two parts. N Engl J Med. 2001;344(14):1058–1066. doi: 10.1056/NEJM200104053441406
