ABSTRACT
Objectives
This study aimed at revealing the underlying mechanisms of the loss and gain of ceftazidime-avibactam susceptibility in a non-carbapenemase-producing hypervirulent Klebsiella pneumoniae (hvKp).
Methods
Here we longitudinally recovered 3 non-carbapenemase-producing K1-ST23 hvKp strains at a one-month interval (KP29105, KP29499 and KP30086) from an elderly male. Antimicrobial susceptibility testing, whole genome sequencing, transcriptomic sequencing, gene cloning, plasmid conjugation, quantitative real-time PCR (qRT-PCR), and SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) were conducted.
Results
Among the 3 hvKp strains, KP29105 was resistant to the third- and fourth-generation cephalosporins, KP29499 acquired resistance to both ceftazidime-avibactam and carbapenems, while KP30086 restored its susceptibility to ceftazidime-avibactam, imipenem and meropenem but retained low-level resistance to ertapenem. KP29105 and KP29499 carried plasmid-encoded genes blaCTX-M-15 and blaCTX-M-71, respectively, but KP30086 lost both. Cloning of gene blaCTX-M-71 and conjugation experiment of blaCTX-M-71-carrying plasmid showed that the transformant and transconjugant were susceptible to ceftazidime-avibactam but had a more than 8-fold increase in MICs. Supplementation with an outer membrane permeabilizer could reduce the MIC of ceftazidime-avibactam by 32 folds, indicating that porins play a key role in ceftazidime-avibactam resistance. The OmpK35 of the 3 isolates was not expressed, and the OmpK36 of KP29499 and KP30086 had a novel amino acid substitution (L359R). SDS-PAGE and qRT-PCR showed that the expression of porin OmpK36 of KP29499 and KP30086 was significantly down-regulated compared with KP29105.
Conclusions
In summary, we reported the rare ceftazidime-avibactam resistance in a non-carbapenemase-producing hvKp strain. Resistance plasmid carrying blaCTX-M-71 and mutated OmpK36 had a synergetic effect on the resistance.
Introduction
Hypervirulent Klebsiella pneumoniae (hvKp) often causes community infections and exhibits high pathogenicity but limited resistance to antibiotics [Citation1]. In one study, the mortality in patients with hvKp bacteraemia was close to 37% [Citation2]. Recently, carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKp), such as K1/K2 hvKp strain, which acquires carbapenem-resistance plasmid, has emerged and poses a new challenge to the treatment of its infection [Citation3–5]. Novel antibiotics are urgently needed to control infections caused by these CR-hvKp isolates.
Ceftazidime-avibactam is a new type β-lactam/β-lactamase inhibitor combination. Avibactam is effective against extended-spectrum β-lactamases (ESBLs), cephalosporinases, serine-carbapenemases and some oxacillinases but not metallo-β-lactamases [Citation6]. Yu F et al. revealed the in vitro activity of ceftazidime-avibactam against CR-hvKp [Citation7]. Unfortunately, although ceftazidime-avibactam-based treatments exhibited initial optimistic efficacy in treating carbapenem-resistant K. pneumoniae (CRKp) infections, some KPC-producing K. pneumoniae isolates might develop resistance, with its mechanisms involving the mutations in blaKPC gene or high expression of the blaKPC gene combined with porins deficiency [Citation8–11]. HvKp resistant to ceftazidime-avibactam was also reported due to producing NDM-type carbapenemase, or the absence of ompK35 and ompK36 genes while producing KPC-2 [Citation12]. However, ceftazidime-avibactam resistance in non-carbapenemase-producing K. pneumoniae isolates has been rarely reported. Both A et al. reported that the mutated CTX-M-14 isoform in K. pneumoniae DT1 exhibited augmented ceftazidime hydrolytic activity and increased the MIC of ceftazidime-avibactam to 32 mg/L [Citation13]. Castanheira M et al. showed that a blaCTX-M-15-carrying K. pneumoniae combined with disrupted OmpK35 could increase the MIC of ceftazidime-avibactam to 4 mg/L, but did not reach the level of resistance [Citation6]. As for non-carbapenemase-producing CR-hvKp isolates, there are almost no such examples.
In this study, we isolated 3 homologous hvKp isolates that do not produce carbapenemases from a chronic obstructive pulmonary disease (COPD) patient, and the second strain was resistant to both ceftazidime-avibactam and carbapenems. This study aimed to reveal the underlying mechanism of the loss and gain of ceftazidime-avibactam susceptibility in the non-carbapenemase-producing hvKp strain.
Materials and methods
Ethics
The use of patient medical records and the K. pneumoniae isolates for research purposes was approved by the Ethics Committee of the China-Japan Friendship Hospital (2022-KY-054).
Clinical data and strain collection
A 92-year-old male had a history of tuberculosis for 70 years, COPD for 10 years and tracheotomy for 8 months. He was hospitalized due to a 3-month cough and expectoration. Three K. pneumoniae isolates (KP29105, KP29499 and KP30086) with different resistant phenotypes were longitudinally recovered from his sputum and bronchoalveolar lavage fluid specimens at a one-month interval. K. pneumonia isolates were identified using matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI-TOFMS, Bruker Daltonik, Germany). After KP29105 was isolated on the 52nd day of admission, meropenem was used continuously for 6 days, and on the 20th day after that, KP29499 was isolated. Meropenem was discontinued for some time, and KP30086 was isolated 10 days after the second use of meropenem. The patient was not administered ceftazidime-avibactam during the observation period and finally died of multiple organ failure and septic shock.
Knockin and knockout experiments
We conducted knockin and knockout experiments among strains KP29499 and KP30086. Specifically, a total of three mutants including strains KP30086+CTX-M-71 (knockin blaCTX-M-71 for KP30086), KP29499-ΔompK36Comp (knockout original mutated ompK36 and knockin wild-type ompK36 for KP29499), and KP29499-ΔCTX-M-71 (knockout blaCT-M-71 from KP29499) were constructed.
The construction of ompK36 and blaCTX-M-71 deletion strains was performed in K. pneumoniae KP29499 using a CRISPR-Cas9-mediated genome-editing method containing pCasKP-apr and pSGKP-spe plasmids [Citation14]. Firstly, the spacers of blaCTX-M-71 and ompk36 were ligated into pSGKP-spe, respectively. Then about 500 bp segment around the deleting sequences was amplificated, and the production and the spacer-introduced pSGKP-spe plasmid were transferred into the L-arabinose-induced pCasKP-apr-harbouring recipient with electrotransformation. The deletion strains KP29499-ΔCTX-M-71 and KP29499-Δompk36 were selected after the cells incubated at 30 °C overnight. Last, the deletion strains were cultured on a sucrose-containing Luria-Bertani (LB) plate at 37 °C for curing the pCasKP-apr and pSGKP-spe plasmid.
The complementation experiment was performed as previously described [Citation15]. The wild-type ompK36 was amplified by PCR using KP29105 as a template. The PCR production and the apramycin-resistant plasmid pDK6 were digested using BamHI and HindIII endonuclease (NEB, USA). Then, T4 DNA ligase (NEB, USA) was used for ligating the vector at 16 °C overnight, and the ligated-plasmid was transformed into KP29499Δompk36 and then selected on a 30 mg/ml apramycin-containing LB plate. The pDK6-ompk36 plasmid-carrying clone was picked up as KP29499-Δompk36Comp. In addition, the knockin of blaCTX-M-71 into strain KP30086 was also carried out using the same method described above. The primers used in the knockin and knockout experiments were listed in Table S1.
Antimicrobial Susceptibility Testing (AST)
AST tests were performed using the microdilution broth method in 96-well plates (MBD, bio-KONT, Ltd. China), mainly including piperacillin, piperacillin-tazobactam, ceftazidime, ceftazidime-avibactam, cefotaxime, cefepime, aztreonam, imipenem, meropenem, and ertapenem. In addition, the MIC of ceftazidime, ceftazidime-avibactam, meropenem and cefotaxime were also tested by adding the outer membrane permeabilizer polymyxin B nonapeptide (PMBN). Briefly, the PMBN mother liquor was prepared, the bacterial solution was adjusted to 0.5 McFarland turbidity, and the bacterial solution and PMBN mother liquor were added to the 96-well antibiotic susceptibility plate sequentially so that the final concentration of PMBN was 10 mg/L.
Hypermucoviscous phenotype determination and the definition of hvKp
The string test was used to identify the hypermucoviscous phenotype of K. pneumoniae strains. Strains exhibiting a viscous string >5 mm were considered hypermucoviscous [Citation16]. HvKp was defined as K. pneumoniae strains with hypermucoviscous phenotype and harbour hypervirulent plasmid in this study.
Whole-Genome Sequencing (WGS) and data analysis
Bacterial genomic DNA was extracted from overnight cultures on Mueller-Hinton broth using TIANGEN Bacterial Genomic DNA Extraction Kit (DP302–02, TIANGEN, Beijing, China) and purified using Agencourt AMPure XP Kit (Beckman Coulter, USA) according to the manufacturer’s instructions. For Illumina sequencing, the genomic DNA libraries were prepared using Nextera XT DNA Library Prep Kit (Illumina, San Diego, USA) according to the manufacturer’s instructions. For nanopore sequencing, a total amount of 500 ng genomic DNA per sample was used as input material for the Oxford Nanopore Technologies (ONT) library preparation, then, all other steps were followed according to the Native barcoding genomic DNA ONT protocol for library preparation and Flongle sequencing. For all three K. pneumoniae strains, WGS was carried out using both the Illumina HiSeq 2500 platform and the nanopore sequencing method on Flongle flow cells to produce short and long reads, respectively. Raw reads from the two sequencing platforms were filtered to remove low-quality sequences and adaptors using skewer [Citation17] and PoreChop (https://github.com/rrwick/Porechop), respectively. The resulting short and long reads were de novo assembled using SPAdes Genome Assembler v3.14.0 [Citation18] and Unicycler v0.4.8 [Citation19] to obtain complete circular genomes. Gene prediction was performed using Prokka 1.12 [Citation20]. PROVEAN [Citation21] was used for predicting the effects of mutation on protein function. The antimicrobial resistance genes, multilocus sequence types (MLST), and plasmid replicons were analyzed via the CGE server (https://cge.cbs.dtu.dk/services/). Virulence genes were identified using the BIGSdb-Kp database provided by the Institut Pasteur (http://bigsdb.web.pasteur.fr). The single nucleotide polymorphism (SNP) was called using Snippy (https://github.com/tseemann/snippy) with KP29105 as a reference. Large insertion/deletion (indel) and rearrangement were analyzed using Mauve software [Citation22]. Insertion sequences were annotated against the ISfinder database [Citation23]. The promoters were predicted using BPROM [Citation24]. The ompK35 and ompK36 genes of KP29105, KP29499 and KP30086 were compared with the corresponding genes of K. pneumoniae NTUH-K2044.
Transcriptomic analysis
The bacteria were inoculated into Mueller-Hinton broth and cultured at 37 °C 200 rpm overnight. The bacterial solution was adjusted to OD600 0.5 and then transferred into a new Mueller-Hinton broth without antibiotics at 1% inoculation rate, and cultured at 37 °C 200 rpm for 3 hours. Bacterial solution (no more than 1 × 109 cells) was taken for total RNA extraction using the enzymatic method (lysozyme) with the RNeasy minikit (Qiagen) following the manufacturer’s instructions. The concentration and quality of RNA were measured by Qubit 3.0. After the extracted RNA was qualified, the ribosomal RNA (rRNA) was removed. Subsequently, the mRNA fragments were randomly interrupted into short fragments by an enzyme digestion method. The library was built in a chain-specific way according to the protocol of the RNA-Seq Library Prep Kit for Illumina (Transgen, Beijing, China). After qualified library inspection, Illumina HiSeq 2500 sequencing was performed to obtain the sequence information of the fragment. FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and skewer [Citation17] were used for quality filtering and adaptor removal, respectively. Transcriptomic analysis was conducted among K. pneumoniae strains KP29105 and KP29499, and their genomes were used as references for their respective RNA-seq paired-end reads. To compare the gene expressions of the two slightly different genomes, a comparative genomic analysis was conducted beforehand. Overall, 5227 genes have one-to-one correspondence in the two genomes. Besides, there were 74 and 75 strain-specific genes in KP29105 and KP29499, respectively. Subread [Citation25] was used for sequence alignment. The software featureCounts [Citation26] was used to tally the expression counts of genes in the reference genome. The differentially expressed genes (DEGs) were analyzed by DESeq2 [Citation27] for the two groups. Principal Component Analysis (PCA) was carried out using the overall expression data among the two groups. KEGG pathway enrichment was performed for the DEGs. Linear discriminant analysis effect size (LEfSe) [Citation28] analysis was performed to determine the features of KEGG pathways.
Cloning and conjugation experiments
The coding sequences of blaCTX-M-15 and blaCTX-M-71 genes and their promoters were cloned into the pGEM-T Easy vector, using the primer pairs in Table S1. The ligation product was then transformed into Escherichia coli DH5α competent cells using heat shock-based transformation method. Conjugation assay was performed according to a previously described method with slight modification [Citation29]. Briefly, both strain K. pneumoniae KP29499 and azide-resistant recipient strain E. coli J53 were cultured overnight on Mueller-Hinton broth and diluted to the 0.5 McFarland standard. Next, 100 μl of donor cells and 400 μl of recipient cells were mixed and inoculated onto a 0.45 µm membrane placed on the surface of a Mueller-Hinton plate at 37 °C overnight. Next, bacteria on the membrane were collected, serially diluted and spread on LB plates containing azide (100 mg/L) and aztreonam (2 mg/L). Antimicrobial susceptibility testing and PCR amplification and sequencing were performed to confirm the successful transfer of resistant plasmid.
Transcriptional analysis by the quantitative real-time PCR (qRT-PCR)
A total of 800 μl of bacterial cultures were used for RNA extraction for each of the 3 K. pneumoniae isolates using the same method as transcriptomic analysis mentioned above. First-strand cDNA was synthesized with 1 µg of total RNA using a RevertAid First strand cDNA synthesis kit (Thermo Fisher Scientific, USA). The transcriptional expression of the genes acrA, acrB, ompK35, ompK36, blaSHV-11 and blaCTX-M was measured using qRT-PCR on an Applied Biosystems QuantStudio 5 real-time PCR system (ThermoFisher Scientific, USA). The amplification conditions were as follows: 50 °C for 2 minutes, 95 °C for 2 minutes, 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. The expression levels of the target genes were normalized to the rpoB gene using ∆∆CT method and using the gene expressions of KP29105 as references. The primers used for qRT-PCR were listed in Table S1. All amplifications were carried out from three different RNA preparations.
Outer membrane protein extraction and analysis
The outer membrane protein fractions were extracted according to the methodology previously described [Citation30] with minor changes. Briefly, 50-ml bacterial cultures were grown overnight in Mueller-Hinton broth. The cells were harvested after being centrifugated at 5000 × g for 10 minutes. The pellets were washed with 1-ml 10 mM N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES), resuspended with 1.5-ml HEPES, and lysed with sonication for 10 seconds 12 times with 10-second interval. Unbroken cells were removed by centrifugation. The supernatants were further centrifuged at 16,500 rpm for 30 minutes at 4 °C. The cell membrane pellets were suspended in 0.2-ml HEPES. The cytoplasmic membranes were solubilized by adding an equal volume of 2% Sarkosyl and incubated at room temperature for 30 minutes with intermittent mixing. The outer membranes were then collected by centrifugation for 30 minutes, washed with 0.5-ml HEPES without resuspension, and suspended in 50-μl HEPES.
The extracted outer membrane proteins were adjusted to the same concentration of 5 mg/ml, mixed with loading buffer, heated at 100 °C for 5 minutes, and then analyzed by denaturing 12% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Outer membrane proteins of K. pneumoniae NTUH-K2044 were used as the positive control to interpret the position of major porins on SDS-PAGE [Citation31]. The protein gels were recovered for subsequent protein identification with liquid chromatography/tandem mass spectrometry (LC-MS/MS).
LC-MS/MS analysis
The protein gel strips were cut into small pieces and decolorized in 50% acetonitrile (ACN)/50 mmol/L NH4HCO3. Eluent was discarded, and the above operations were repeated until the gel strips became colourless. 100% ACN was added and dehydrated for 30 minutes. ACN was discarded and the gel pieces were dried at room temperature. Trypsin was added and incubated at 4 °C for 40 minutes, and then digested in 37 °C water bath for 16 hours. Extract solution (5% TFA-50% ACN-45% water) was added to extract peptide. The resulting peptide supernatant was transferred to a new tube, desalted, lyophilized and resuspended in 0.1% formic acid and 2% ACN for LC-MS/MS analysis. After that, the processed samples were analyzed by LC-MS/MS to obtain the raw files of the original results of mass spectrometry. The raw files were analyzed by using the software MaxQuant (1.6.2.10) to obtain the identification results.
Growth kinetics
Growth curves for KP29105, KP29499 and KP30086 strains were measured as follows: bacterial cultures grown to exponential phase at Mueller-Hinton broth were adjusted to OD600 0.09–0.10, and diluted 1:100. Three aliquots of 200 μl per dilution were transferred into a 96-well microtiter plate and covered with paraffin oil. Samples were incubated at 37 °C and shaken before measurement in a Multiskan GO Microplate Reader (Thermo Fisher Scientific, USA). All experiments were performed in triplicate.
Statistical analysis
Statistically significant differences in growth kinetics were tested using the R package “statmod” [Citation32]. Student’s t-test was employed to check the expression difference between the two groups. A P-value of 0.05 or less was defined as significant.
Results
Homology and plasmid profiles
Taking K. pneumoniae strain KP29105 as a reference, a total of 13 SNPs were found in strains KP29499 and KP30086, of which 5 mutations were located in the non-coding regions and 8 mutations were located in the coding regions (). In addition, there were 7 and 12 SNPs for strains KP29499 and KP30086, respectively.
Table 1. Resistance and virulence genes in three K. pneumoniae strains and mutations in the coding region of strains KP29499 and KP30086 using KP29105 as reference.
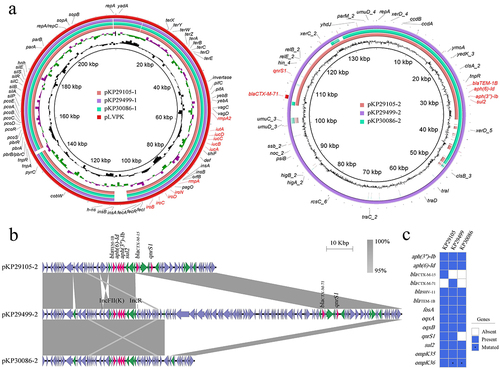
All the 3 K. pneumoniae strains were hypermucoviscous with the positive string tests. Moreover, they belonged to ST23 and had the capsular type of K1. Plasmid analysis indicated that they all contained 2 plasmids (pKP29105–1 (CP091062), pKP29105–2 (CP091063), pKP29499–1 (CP089599), pKP29499–2 (CP089600), pKP30086–1 (CP091059), pKP30086–2 (CP091060)). The larger one was a pLVPK-like hypervirulent plasmid (IncHI1B(pNDM-MAR) and repB replicons), harbouring rmpA/rmpA2 genes, iucABCDiutA and iroBCDN virulence clusters ( and ). Therefore, they all belonged to hvKp according to our definition. The smaller plasmid (IncFII(K) replicon) was a multi-resistance plasmid, showing different structures among the 3 isolates. Namely, pKP29499–2 was a hybrid plasmid, with an IncR type plasmid integrated into pKP29105–2 behind an insertion sequence IS6, whereas pKP30086–2 had a large fragment deletion compared with pKP29499–2 ().
Figure 1. Genomic profiles of these 3 clinical K. pneumoniae strains KP29105, KP29499 and KP30086. (a) Circular maps of plasmids in these 3 strains (b) Major structural features of the multi-resistance plasmids in the 3 K. pneumoniae isolates. pKP29499–2 integrated an IncR type plasmid after an insertion sequence IS6 of pKP29105–2, and blaCTX-M-15 becomes blaCTX-M-71 by single amino acid substitution. Whereas pKP30086–2 had a large fragment deletion compared with pKP29499–2, and the blaCTX-M gene was located in this fragment (figure 1B). Boxed arrows indicate the positions of open reading frames (ORFs) and their directions of transcription. ORFs encoding resistance genes are portrayed by red arrows. Green arrows indicate insertion sequences. Gray areas denote more than 95% DNA identity between sequences. (c) Resistance genes in the 3 clinical K. pneumoniae strains. Asterisks represent genes with mutations.
Antimicrobial resistance profiles
The carbapenem-susceptible K. pneumoniae strain KP29105 was first isolated on the 52nd day of admission, after which meropenem was administrated for several days; a month later, KP29499 was isolated and acquired resistance to carbapenem and ceftazidime-avibactam; another month later, KP30086 was isolated, which lost the resistant phenotype to ceftazidime-avibactam, carbapenems, ceftazidime, cefotaxime, cefepime and aztreonam, but the MICs of carbapenems increased 2–4 times compared with those of strain KP29105 ().
Table 2. Antibiotic resistance characteristics (MICs, mg/L) of 3 clinical K. pneumoniae isolates and the transconjugant, transformants and mutants.
The resistance genes were analyzed. As shown in , all the 3 hvKp isolates carried the plasmid-encoded blaTEM-1B gene and chromosomal blaSHV-11 gene. Notably, KP29105 had a blaCTX-M-15 gene, KP29499 harboured a blaCTX-M-71 gene with a single-base mutation (G721T, resulting in Gly241Cys substitution) compared to blaCTX-M-15, and KP30086 lost the blaCTX-M gene.
We cloned the blaCTX-M-15 and blaCTX-M-71 genes into pGEM-T Easy vectors and transformed them into E. coli DH5α competent cells. AST showed that the transformants DH5α (CTX-M-15) and DH5α (CTX-M-71) obtained resistance to cephalosporins and aztreonam but not piperacillin-tazobactam, ceftazidime-avibactam and carbapenems (). The MIC of ceftazidime-avibactam for the transformant DH5α (CTX-M-71) was 1 mg/L, which was 4- and 8-fold that of DH5α (CTX-M-15) and the recipient E. coli DH5α, respectively. In addition, the plasmid pKP29499–2 was successfully transferred into the recipient E. coli J53, and the transconjugant exhibited the same resistance phenotype as the transformants but with different MICs (). Notably, the transconjugant also had a MIC of 1 mg/L for ceftazidime-avibactam.
After co-treatment with PMBN at the concentration of 10 mg/L, the MIC of ceftazidime, ceftazidime-avibactam, cefotaxime and meropenem decreased 4 to 32 folds, as shown in .
Table 3. Antibiotic resistance characteristics (MICs, mg/L) of K. pneumoniae strain KP29499 to ceftazidime, ceftazidime-avibactam, meropenem and ceftriaxone.
The AST results for the three knockin and knockout strains were showed in . As expected, strain KP30086+CTX-M-71 recovered resistance to ceftazidime, ceftazidime-avibactam, cefotaxime, cefepime, aztreonam, meropenem and ertapenem. Strain KP29499-ΔompK36Comp showed decreased MIC to ceftazidime-avibactam (from 32 to 4 mg/L), meropenem (from 16 to 8 mg/L) and ertapenem (from 64 to 16 mg/L). Strain KP29499-ΔCTX-M-71 became susceptible to ceftazidime-avibactam, cefotaxime, cefepime, aztreonam, meropenem and imipenem.
Outer membrane proteins and efflux pumps profiling
The ompK35 and ompK36 genes among the 3 hvKp strains were analyzed with K. pneumoniae NTUH-K2044 as a reference. No indels and rearrangements were detected around ompK35 and ompK36. No mutations were found in the ompK35 gene and its promoter region. A single-base mutation (T1076G) was found in the ompK36 gene in both strains KP29499 and KP30086. The mutation led to amino acid substitution (L359R) and had a deleterious effect on the function of OmpK36 according to PROVEAN.
The expression at the transcriptional level of ompK35 and ompK36 genes was analyzed by qRT-PCR. The ompK35 expression was deficient in all the 3 K. pneumoniae strains. In addition, strains KP29499 and KP30086 had a significantly lower expression for the ompK36 gene compared with KP29105 ().
Figure 2. Porin profiles and growth curves of K. pneumoniae strains KP29105, KP29499 and KP30086. (a) Relative expression at the transcriptional level of genes ompK35, ompK36 and blaCTX-M. Data were normalized to the expression levels of gene rpoB. K. pneumoniae NTUH-K2044 and KP29105 were set as the basic expression for porins and CTX-M, respectively. (b) SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) of outer membrane proteins. K. pneumoniae NTUH-K2044 was used as reference. (c) Growth curves of the K. pneumoniae isolates. Three independent experiments were carried out. The significance level was as follows: *p < 0.05.
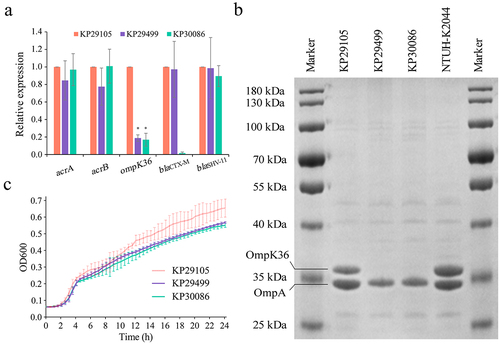
The expression of porins at the translational level was tested using SDS-PAGE, and the identification of proteins was carried out using LC-MS/MS. The results showed that OmpA could be found in all the 3 isolates, OmpK36 was lost from K. pneumoniae strains KP29499 and KP30086, and OmpK35 was absent in all the 3 isolates ().
The expression of genes acrA and acrB was examined using the qRT-PCR method. No significant difference was observed among the 3 strains ().
Transcriptomic data for strains KP29105 and KP29499
Transcriptomic analysis was performed to detect the potential mechanisms involved in the resistance of ceftazidime-avibactam and carbapenems in strain KP29499. A total of 675 DEGs were identified. Among them, 359 up-regulated and 316 down-regulated DEGs were observed after data filtering when compared to strain KP29105. PCA analysis showed that the gene expressions of strains KP29105 and KP29499 were clustered into two distinct groups (Figure S1A). The results of KEGG enrichment analysis were showed in Figure S1B-C.
The expression levels of genes related to porins, efflux pumps and β-lactamases were examined. Similar to the qRT-PCR results, the outer membrane gene ompK35 was expressed at a very low level among strains KP29499 and KP29105. The ompA expression of the two strains was very high, and there was no significant difference. In addition, lower expression of gene ompK36 was observed in strain KP29499 but not to a statistically significant level. Moreover, there were no significant differences in the expression of β-lactamase genes blaCTX-M, blaTEM, blaSHV, and efflux pump-related genes such as eefAB, mdtABC, acrAB, acrR, marR, oqxR, ramR, rarA, rpsJ and soxR.
Growth curves
The 24-hour growth curves of the K. pneumoniae strains KP29105, KP29499 and KP30086 were tested. KP29105 had a higher growth rate compared to the other strains (), but the difference was not statistically significant.
Discussion
Ceftazidime-avibactam provides a novel therapeutic option against CRKp strains [Citation33], but the resistance strains have been reported worldwide [Citation34,Citation35]. Ceftazidime-avibactam resistance is usually caused by blaKPC-related mechanisms [Citation36]. In this study, we reported the ceftazidime-avibactam resistance in a non-carbapenemase-producing hvKp strain KP29499, which has been rarely reported. Most notably, this strain is a K1-ST23 hvKp strain, which is worrisome and should be monitored closely.
Strain KP29499 contained 3 β-lactamase genes: blaTEM-1B, blaSHV-11 and blaCTX-M-71. TEM-1B confers resistance to penicillins and narrow-spectrum cephalosporins [Citation37]. SHV-11 is not ESBL, and it is intrinsic in Klebsiella and had a low level of expression according to the transcriptomic analysis, which was consistent with previous reports [Citation38,Citation39]. Therefore, only the functions of CTX-M-71 were examined by cloning and transformation experiments.
Strains KP29105 and KP29499 carried blaCTX-M-15 and blaCTX-M-71 genes, respectively, while KP30086 lost both. There is no consensus on the hydrolytic activity of CTX-M-71 and CTX-M-15 for third- and fourth-generation cephalosporins [Citation40,Citation41]. Our study showed that the transformant DH5α (CTX-M-71) had the same MICs for ceftazidime and cefotaxime compared to DH5α (CTX-M-15), and higher MICs for cefepime, aztreonam and ceftazidime-avibactam. Notably, although neither the transformants nor the transconjugant acquired resistance to ceftazidime-avibactam, a more than 2-fold increase in MICs from ≤ 0.125 to 0.25 or 1 was observed, indicating that the blaCTX-M gene and other mechanisms were involved in the ceftazidime-avibactam resistance.
PMBN (10 mg/L), an outer membrane permeabilizer, can significantly decrease the MICs of ceftazidime and ceftazidime-avibactam by 32 folds, indicating that the porins played an important role in the ceftazidime-avibactam resistance, as reported previously [Citation6,Citation42]. OmpK35 and OmpK36 are two main non-specific porins in K. pneumoniae, permitting the diffusion of some nutrients and hydrophilic molecules such as β-lactams into the cell [Citation43]. The mutation, deficiency, or loss of function of OmpK36 or OmpK35 porins, combined with KPC or CTX-M might result in the elevated MIC of ceftazidime-avibactam [Citation6,Citation8,Citation44,Citation45]. In our study, 3 clinical hvKp isolates shared the same ompK35 gene as the typical hvKp NTUH-K2044, and KP29105 had the wild-type ompK36 gene, but KP29499 and KP30086 had a novel amino acid substitution (L359R) for ompK36, which had a deleterious effect on the protein function according to PROVEAN result.
The porins OmpK35 and OmpK36 were then analyzed at the transcriptional and translational levels. For porin OmpK35, all the 3 strains had low expression at the transcriptional level and were not visible at the translational level. According to previous reports, K. pneumoniae NTUH-K2044 expresses OmpK36 and lacks OmpK35 [Citation46], the same result was obtained in our study. In addition, strains KP29499 and KP30086 had significantly lower transcriptional levels for gene ompK36 compared with KP29105. Meanwhile, the SDS-PAGE experiment showed that OmpK36 was absent from KP29499 and KP30086 at the translational level. A similar phenomenon has been previously reported in phosphate-regulated porin PhoE, in which another unknown protein involved in promoter recognition has been mutated, causing phoE to fail to be transcribed/translated correctly [Citation47]. There could be other mechanisms resulting in the invisibility of OmpK36 at the translational level, including lower expression level, low quantity of this protein or its constructional alteration by post-translational modifications [Citation48].
Although strain KP30086 lost the porin OmpK36, it didn’t obtain the resistance for ceftazidime-avibactam, indicating that the OmpK36 mutant alone cannot cause ceftazidime-avibactam resistance. Strain KP29499, the MIC of ceftazidime-avibactam was 32 mg/L, had a CTX-M-type ESBL and deficient OmpK36, and the MIC was reduced to 1 mg/L after PMBN was added, which verified that the high ceftazidime-avibactam MICs in KP29499 was associated with the co-existence of the mutated OmpK36 and CTX-M-71. Besides, the MICs of ceftazidime-avibactam of the newly constructed strains KP30086+CTX-M-71 and KP29499-ΔCTX-M-71 demonstrated the role of blaCTX-M-71 in ceftazidime-avibactam resistance, and strain KP29499-ΔompK36Comp revealed the role of ompK36 in ceftazidime-avibactam resistance.
OmpK36 is the primary channel for the permeation of ertapenem and meropenem but not for imipenem [Citation49]. Mutations in the ompK36 gene confer high-level carbapenem resistance to KPC-producing K. pneumoniae strains [Citation50]. OmpK36 deficiency in CTX-M-15-producing K. pneumoniae can lead to resistance to ertapenem and reduce susceptibility to meropenem [Citation43,Citation49]. Herein, a consistent result was obtained. Our results showed that in strain KP30086, OmpK36 deficiency alone can cause low-level ertapenem resistance and an increase in MICs of meropenem but not to a resistance level. Moreover, the co-existence of CTX-M-71 and OmpK36 deficiency in KP29499 caused the resistance of the 3 carbapenems, with the highest MIC in ertapenem, followed by meropenem and imipenem. Before the CRKp strain KP29499 was recovered from the patient, meropenem was administrated for several days, which may relate to the mutation in the ompK36 gene, as suggested by the previous studies [Citation49,Citation51]. OmpK36 absence is not only associated with antibiotic resistance but also decreases bacterial fitness [Citation43,Citation52]. The same phenomenon was observed in the 3 K. pneumoniae strains, namely, KP29499 and KP30086 had relatively lower growth rates compared with KP29105.
In summary, the within-host evolution of ceftazidime-avibactam resistance in K. pneumoniae is far from being fully elucidated. In the present study, the repeated in-vivo loss and gain of the ceftazidime-avibactam susceptibility in a series of clinical hvKp strains were captured, with CTX-M-71 and mutated OmpK36 involved in ceftazidime-avibactam resistance. Close attention is required to prevent the spread of these K. pneumoniae strains.
Acknowledgements
We would like to thank all our colleagues in the Laboratory of Clinical Microbiology and Infectious Diseases for their assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Supplemental data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2024.2348251
Additional information
Funding
References
- Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–12. doi: 10.1128/AAC.01127-16
- Li J, Ren J, Wang W, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(4):679–689. doi: 10.1007/s10096-017-3160-z
- Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi: 10.1093/jac/dkz446
- Zhao J, Liu C, Liu Y, et al. Genomic characteristics of clinically important ST11 Klebsiella pneumoniae strains worldwide. J Glob Antimicrob Resist. 2020;22:519–526. doi: 10.1016/j.jgar.2020.03.023
- Tian D, Liu X, Chen W, et al. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect. 2022;11(1):1936–1949. doi: 10.1080/22221751.2022.2103454
- Castanheira M, Doyle TB, Hubler C, et al. Ceftazidime-avibactam activity against a challenge set of carbapenem-resistant enterobacterales: Ompk36 L3 alterations and β-lactamases with ceftazidime hydrolytic activity lead to elevated MIC values. Int J Antimicrob Agents. 2020;56(1):106011. doi: 10.1016/j.ijantimicag.2020.106011
- Yu F, Lv J, Niu S, et al. In vitro activity of ceftazidime-avibactam against carbapenem-resistant and hypervirulent Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. 2018;62(8):e01031–18. doi: 10.1128/AAC.01031-18
- Gaibani P, Re MC, Campoli C, et al. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: epidemiology and genomic characterization. Clin Microbiol Infect. 2020;26(4):.e516.1–.e516.4. doi: 10.1016/j.cmi.2019.11.011
- Wang C, Zhao J, Liu Z, et al. In vivo Selection of Imipenem Resistance Among Ceftazidime-Avibactam-Resistant, Imipenem-Susceptible Klebsiella pneumoniae Isolate With KPC-33 Carbapenemase. Front Microbiol. 2021;12:727946. doi: 10.3389/fmicb.2021.727946
- Venditti C, Butera O, Meledandri M, et al. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin Microbiol Infect. 2021;27(7):.e1040.1–.e1040.6. doi: 10.1016/j.cmi.2021.03.001
- Liu L, Feng Y, Long H, et al. Sequence type 273 carbapenem-resistant Klebsiella pneumoniae carrying blaNDM-1 and blaIMP-4. Antimicrob Agents Chemother. 2018;62(6). doi: 10.1128/AAC.00160-18
- Li D, Liao W, Huang HH, et al. Emergence of hypervirulent Ceftazidime/Avibactam-resistant Klebsiella pneumoniae isolates in a Chinese tertiary hospital. Infect Drug Resist 13:2673–2680. 10.2147/IDR.S257477. 2020.
- Both A, Buttner H, Huang J, et al. Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J Antimicrob Chemother. 2017;72(9):2483–2488. doi: 10.1093/jac/dkx179
- Wang Y, Wang S, Chen W, et al. CRISPR-Cas9 and CRISPR-Assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl Environ Microbiol. 2018;84(23):e01834–18. doi: 10.1128/AEM.01834-18
- Li Z, Liu X, Lei Z, et al. Genetic diversity of polymyxin-resistance mechanisms in clinical isolates of carbapenem-resistant Klebsiella pneumoniae: a multicenter study in China. Microbiol Spectr. 2023;11(2):e0523122. doi: 10.1128/spectrum.05231-22
- Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718
- Jiang H, Lei R, Ding SW, et al. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf. 2014;15(1):182. doi: 10.1186/1471-2105-15-182
- Prjibelski A, Antipov D, Meleshko D, et al. Using SPAdes De Novo Assembler. Curr Protoc Bioinform. 2020;70(1):e102. doi: 10.1002/cpbi.102
- Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153
- Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLOS ONE. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688
- Darling AC, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704
- Siguier P, Perochon J, Lestrade L, et al. Isfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(90001):D32–6. doi: 10.1093/nar/gkj014
- Solovyev V, Salamov A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. Metagenomics And Its Applications In Agriculture. Hauppauge (NY): Nova Science Publishers; 2011. p. 61–78.
- Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60
- Yang X, Wai-Chi Chan E, Zhang R, et al. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019;4(12):2039–2043. doi: 10.1038/s41564-019-0566-7
- Carlone GM, Thomas ML, Rumschlag HS, et al. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24(3):330–332. doi: 10.1128/jcm.24.3.330-332.1986
- Dupont H, Gaillot O, Goetgheluck A-S, et al. Molecular characterization of carbapenem-nonsusceptible enterobacterial isolates collected during a prospective interregional survey in France and susceptibility to the novel ceftazidime-avibactam and aztreonam-avibactam combinations. Antimicrob Agents Chemother. 2016;60(1):215–221. doi: 10.1128/AAC.01559-15
- Giner G, Smyth GK. Statmod: probability calculations for the inverse gaussian distribution. R J. 2016;8(1):339–351. doi: 10.32614/RJ-2016-024
- Yin D, Wu S, Yang Y, et al. Results from the China Antimicrobial Surveillance Network (CHINET) in 2017 of the in vitro activities of ceftazidime-avibactam and ceftolozane-tazobactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63(4):e02431–18. doi: 10.1128/AAC.02431-18
- Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):.e124.1–.e124.4. doi: 10.1016/j.cmi.2019.08.020
- Humphries RM, Hemarajata P. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother. 2017;61(6):00537–17. doi: 10.1128/AAC.00537-17
- Xu M, Zhao J, Xu L, et al. Emergence of transferable ceftazidime–avibactam resistance in KPC-producing Klebsiella pneumoniae due to a novel CMY AmpC β-lactamase in China. Clin Microbiol Infect. 2022;28(1):.e136.1–.e136.6. doi: 10.1016/j.cmi.2021.05.026
- Salverda ML, De Visser JA, Barlow M. Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34(6):1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x
- Rice LB, Carias LL, Hujer AM, et al. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin- tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44(2):362–367. doi: 10.1128/AAC.44.2.362-367.2000
- Nuesch-Inderbinen MT, Kayser FH, Hachler H. Survey and molecular genetics of SHV beta-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41(5):943–949. doi: 10.1128/AAC.41.5.943
- Schneider I, Queenan AM, Markovska R, et al. New variant of CTX-M-Type extended-spectrum β-lactamases, CTX-M-71, with a Gly238Cys substitution in a K lebsiella pneumoniae isolate from Bulgaria. Antimicrob Agents Chemother. 2009;53(10):4518–4521. doi: 10.1128/AAC.00461-09
- Sabatini A, Brisdelli F, Celenza G, et al. Interaction of carbapenems and β-lactamase inhibitors towards CTX-M-15 and CTX-M-15 G238C mutant. J Glob Antimicrob Resist. 2017;10:95–100. doi: 10.1016/j.jgar.2017.04.004
- Pages JM, Peslier S, Keating TA, et al. Role of the outer membrane and Porins in susceptibility of β-lactamase-producing Enterobacteriaceae to ceftazidime-avibactam. Antimicrob Agents Chemother. 2015;60(3):1349–1359. doi: 10.1128/AAC.01585-15
- Fajardo-Lubian A, Ben Zakour NL, Agyekum A, et al. Host adaptation and convergent evolution increases antibiotic resistance without loss of virulence in a major human pathogen. PLOS Pathog. 2019;15(3):e1007218. doi: 10.1371/journal.ppat.1007218
- Shen Z, Ding B, Ye M, et al. High ceftazidime hydrolysis activity and porin OmpK35 deficiency contribute to the decreased susceptibility to ceftazidime/avibactam in KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2017;72(7):1930–1936. doi: 10.1093/jac/dkx066
- Nelson K, Hemarajata P, Sun D, et al. Resistance tO ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother. 2017;61(10):e00989–17. doi: 10.1128/AAC.00989-17
- Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M, et al. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2018;52(6):898–905. doi: 10.1016/j.ijantimicag.2018.03.020
- Kaczmarek FM, Dib-Hajj F, Shang W, et al. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla ACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother. 2006;50(10):3396–3406. doi: 10.1128/AAC.00285-06
- Foudraine DE, Strepis N, Stingl C, et al. Exploring antimicrobial resistance to beta-lactams, aminoglycosides and fluoroquinolones in E. coli and K. pneumoniae using proteogenomics. Sci Rep. 2021;11(1):12472. doi: 10.1038/s41598-021-91905-w
- Findlay J, Hamouda A, Dancer SJ, et al. Rapid acquisition of decreased carbapenem susceptibility in a strain of Klebsiella pneumoniae arising during meropenem therapy. Clin Microbiol Infect. 2012;18(2):140–146. doi: 10.1111/j.1469-0691.2011.03515.x
- Shields RK, Clancy CJ, Hao B, et al. Effects of Klebsiella pneumoniae carbapenemase subtypes, extended-spectrum β-lactamases, and porin mutations on the in vitro activity of ceftazidime-avibactam against carbapenem-resistant K. pneumoniae. Antimicrob Agents Chemother. 2015;59(9):5793–5797. doi: 10.1128/AAC.00548-15
- Grobner S, Linke D, Schutz W, et al. Emergence of carbapenem-non-susceptible extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J Med Microbiol. 2009;58(7):912–922. doi: 10.1099/jmm.0.005850-0
- Rocker A, Lacey JA, Belousoff MJ, et al. Global Trends in Proteome Remodeling of the Outer Membrane Modulate Antimicrobial Permeability in Klebsiella pneumoniae. MBio. 2020;11(2):e00603–20. doi: 10.1128/mBio.00603-20

