ABSTRACT
Drosophila Jumonji/Jarid2 (dJmj) has been identified as a component of Polycomb repressive complex 2. However, it is suggested that dJmj has both PRC-dependent and –independent roles. Subcellular localization of dJmj during spermatogenesis is unknown. We therefore performed immunocytochemical analyses with specific antibodies to dJmj and tri-methylation at lysine 27 on histone H3 (H3K27me3). Interestingly, dJmj exclusively localizes at nucleolus in the late growth stage. Examination of the dJmj localization in various Polycomb group (PcG) mutant lines at the late growth stage allowed identification of some PcG genes, including Polycomb (Pc), to be responsible for dJmj recruitment to nucleolus. In addition, we found that size of nucleolus was decreased in some of these mutant lines. In a mutant of testis-specific TAF homolog (tTAF) that is responsible for nucleolus localization of Pc, dJmj signals were detected not only at nucleolus but also on the condensed chromatin in the late growth stage. Duolink In situ Proximity ligation assay clarified that Pc interacts with dJmj at nucleolus in the late growth stage. Furthermore, the level of H3K27me3 decreased in nuclei at this stage. Taken together, we conclude that tTAF is responsible for recruitments of dJmj to nucleolus in the late growth stage that appears to be mediated by Pc. Compartmentalization of dJmj in nucleolus together with some of PcG may be necessary to de-repress the expression of genes required to cellular growth and proliferation in the following meiotic divisions.
Introduction
The JmjC domain was initially characterized as a conserved domain among jumonji (Jmj) family proteins, including Jumonji/Jarid2 (Jmj), RBP2 and SMCX, and has subsequently been identified in more than 100 proteins in prokaryotic and eukaryotic organisms.Citation1-3 JmjC-containing proteins have been shown to play important roles in various biological processes, including cellular differentiation, DNA repair and regulation of heterochromatin.Citation4-6 These JmjC-containing proteins are considered to regulate chromatin and/or transcription. Jmj contains a DNA-binding domain, denoted as the AT-rich interaction domain (ARID), a zinc finger domain, a jumonji N (JmjN) domain and a JmjC domain.Citation7,8 Unlike the other members of the Jmj family proteins, Jmj has a long N-terminal moiety devoid of characterized domains. Although many JmjC domain-containing proteins have been shown to catalyze lysine demethylation, the residues required for iron and ascorbate binding, essential for demethylation activity, are not conserved in the JmjC domain of Drosophila Jumonji (dJmj).Citation8 Therefore, despite of having a JmjC-domain, dJmj has been predicted and reported, to be catalytically inactive as a histone demethylase. Studies with imaginal discs and polytene chromosomes in Drosophila revealed that dJmj is required to fine-tune global tri-methylation at lysine 27 on histone H3 (H3K27me3) levels.Citation9 Even if dJmj would be inactive as a histone demethylase, it might still be able to bind to chromatin and prevent spreading of the H3K27me3 mark, such as opposing the positive spreading effect of Extra sexcombs (Esc).Citation10
Trithorax and Polycomb group (PcG) genes play antagonistic roles in determining whether a gene is transcriptionally turned on or off respectively.Citation11 In Drosophila, so far 4 distinct complexes: pleiohomeotic repressive complex 1 (PhoRC), Polycomb repressive complex 2 (PRC2), Polycomb repressive complex 1 (PRC1), and also Polycomb repressive deubiquitinase (PRDUB), have been described to play a role in PcG-mediated gene repression.Citation12 Recently, Jmj has been characterized as a component of PRC2 in mammalian embryonic stem (ES) cells.Citation13 Although dJmj has also been identified as a component of PRC2 in Drosophila, gene expression and Chromatin immunoprecipitation (ChIP)-sequence analyses suggest both PRC-dependent and –independent role for dJmj.Citation9
Although it is reported that dJmj plays a role in metamorphosis, contribution of dJmj in Drosophila male spermatogenesis remains to be clarified. During Drosophila spermatogenesis, germ stem cells divide asymmetrically, producing a differentiating daughter cell (spermatogonium) and a self-renewing stem cell.Citation14 Each spermatogonium undergoes mitotic division 4 times and generates 16 spermatocytes. During division, spermatogonia are connected each other by ring channels. Sixteen spermatocytes derived from 4 rounds of mitosis generate a cell unit called a cyst and they undergo a cell growth before 2 consecutive meiotic divisions, meiosis I and meiosis II. This stage is designated as a growth stage. Then the nuclei condense and the mitochondria form the characteristic structure called Nebenkern. These appearance looks like an onion and therefore this stage is called onion stage. Then, the flagellar axonema elongates. The cystic bulge is pushed backward after elongation, sweeping the cytoplasm and organelles out of the sperm flagella. This stage is called elongation stage. Then the heads of spermatids containing nuclei elongate and condense more by replacing histones with protamines. This stage is called canoe stage and thereafter spermatogenesis completes.Citation15,16
Previous study indicated that several components of PRC1 such as Polycomb (Pc), Polyhomeotic (Ph) and Drosophila Ring protein (dRing) are co-localized to the same nucleolar subcompartment with testis-specific TATA-binding protein associated factors (tTAFs) in Drosophila spermatocytes.Citation17 However, other studies indicate that Pc and tTAFs associate with chromosome masses and also accumulate in clumps at the periphery of the nucleolus or coat the nucleolar surface appearing as a ring around the nucleolus in the spermatocytes.Citation18 On the other hand, subcellular localization of Jmj during spermatogenesis is unknown both in mammals and Drosophila. In the present study, we performed immunocytochemical analyses with a specific antibody to dJmj to examine subcellular localization of dJmj during spermatogenesis in wild-type flies. Subsequently, we focused on the characteristic nucleolus localization of the dJmj signal in the late growth stage and examined the localization of dJmj in mutant lines of PcG genes or tTAF.
Results
Anti-dJmj immunofluorescence analysis of wild-type testes during spermatogenesis and spermiogenesis
To examine subcellular localization of dJmj during spermatogenesis and spermiogenesis, we immunostained wild-type male testes with the affinity purified anti-dJmj IgG (). This antibody specifically detects the 252 kDa dJmj protein in the Western immunoblot analysis with adult male whole extracts.Citation19 Phase contrast image allows us to identify nucleus in the growth stage, and Nebenkern in the onion stage. Under phase contrast, a prominent dark nucleolus can be easily identified in each wild-type primary spermatocyte (). This was further confirmed by immunostaining with the anti-Fibrillarin antibody that was commonly used as a nucleolus marker.Citation20 The Fibrillarin signal clearly merged with the dark structure under phase contrast (). 4′, 6-diamidino-2-phenylindole (DAPI) staining also confirmed the nucleus () or the condensed heterochromatin in nucleus at the early and late growth stages ().
Figure 1. Anti-dJmj and H3K27me3 immunofluorescence analyses in wild-type Drosophila testes from spermatogonium to late growth stage. (A, G, M) Staining of spermatocytes with DAPI. (B, H, N, N″) Immunostaining of testes with the anti-dJmj antibody. (E, K, Q, Q″) Immunostaining of testes with the anti-H3K27me3 antibody. (M′″) Immunostaining of testes with the anti-Fibrillarin antibody. (D, J, P, P,″ N″′) Phase contrast images of spermatocytes. (C, I, O, O″) Merged DAPI staining with anti-dJmj staining images are shown. (F, L, R, R″) Merged DAPI staining with anti-H3K27me3 staining images are shown. (O″′) Merged anti-Fiblirallin staining with Phase contrast images are shown. (A-F) spermatogonium, (G-L) early growth stage, (M-R, M″–R,″ M″′–O″′) late growth stage. (A′–R′) Magnified images of each stage. (M″–R″) Immunostaining of spermatocytes in the early growth stage without 1st antibody. (M′–R′, M″′–O″′) A nucleus is encircled by solid line and a nucleolus is by dashed line. (K, L), (Q, R) the other cyst from (G-J), (M-P). Scale bars = 50 μm (G-R, M″–R″), 10 μm (A-F), 0.1μm (A′–F′, G′–R′, M″′–O′″).
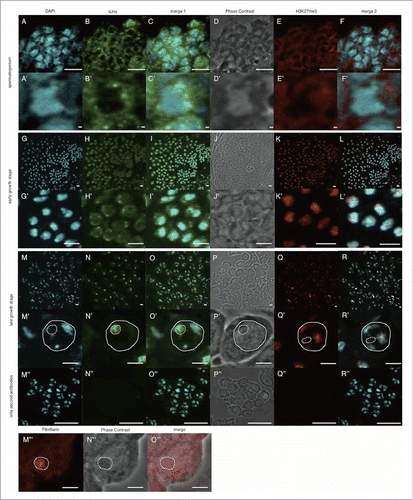
In the spermatogonium dJmj signals were mainly detected in perinuclear region of the cytoplasm (). In the early growth stage, dJmj signals characteristically appeared as dotted signals in nuclei and some of them located at periphery of the condensed chromatin in nuclei (). Interestingly, at the following late growth stage, dJmj signals accumulated in nucleolus () with weak signal in the nucleoplasm and no detectable signal in the cytoplasm. No such dJmj signal was detectable in the control experiments with the second antibody only (). In the next onion stage, dJmj signals appeared to be mainly distributed in nucleus (). dJmj signal was no more detectable in both cytoplasm and nuclei at the elongation stage () and the canoe stage (). These results suggest that dJmj may have a role in early stage of spermatogenesis and/or before the spermatocyte growth stage in spermatogonial stem cell and/or spermatogonial cell amplification division stages.
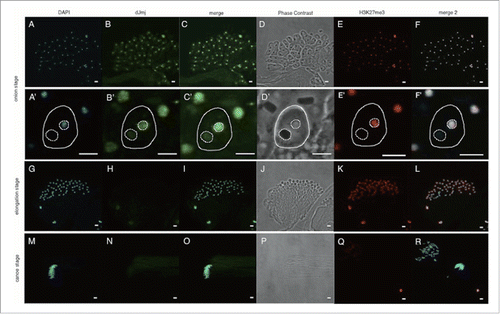
Figure 2. Anti-dJmj and H3K27me3 immunofluorescence analyses in wild-type Drosophila testes from onion stage to canoe stage. (A, G, M) Staining of spermatids with DAPI. (B, H, N) Immunostaining of testes with the anti-dJmj antibody. (E, K, Q) Immunostaining of testes with the anti-H3K27me3 antibody. (D, J, P) Phase contrast images of spermatocytes. (C, I, O) Merged DAPI staining with anti-dJmj staining images are shown. (F, L, R) Merged DAPI staining with anti-H3K27me3 staining images are shown. (A-F) onion stage, (G-L) elongation stage, (M-R) canoe stage. (A′–R′) Magnified images of each stage. (A′–F′) A spermatid is encircled by solid line. A nucleus is encircled by right dashed line and a nebenkern is by left dashed line. (Q, R) the other cyst from (M-P). Scale bars = 50 μm (A-R), 0.1 μm (A′–F′).
Localization pattern of dJmj signals in the late growth stage in PcG mutant lines
We focused on nucleolus localization of dJmj signal in the late growth stage in the wild-type spermatocyte (). Previous report showed that Jmj is a component of PRC2 in mouse ES cells.Citation13 In Drosophila, dJmj has also been identified as a component of PRC2.Citation3 Moreover Pc, the component of PRC1, is reported to localize at the nucleolus in the late growth stage.Citation17 We confirmed Pc localization in Drosophila spermatocytes by immunocytochemical analyses. Immunostaining of testes from Pc-GFP line with the anti-GFP antibody showed that in the early growth stage, Pc signals were detected at condensed chromatin (Fig. S1 A-D′) in contrast to perinuclear localization of dJmj (). However, in late growth stage, similar to dJmj, Pc signals were mainly concentrated at the nucleolus with weak signals throughout nucleus (Fig. S1 E–H′). Based on these observations, we examined the dJmj signal localization in various PcG mutant lines and found that localization pattern of dJmj signals significantly changes in some mutant lines ().
The observed localization pattern of dJmj signals could be divided into 3 groups. In the first group, dJmj localized at the nucleolus similarly to the wild-type spermatocyte (). In the second group, dJmj signal localized at the whole nuclei (). In the third group, the intensity of dJmj signals was different from that of wild-type (). In the Esc, Sex comb on midleg (Scm), Posterior sex combs (Psc) and polyhomeotic distal (ph-d) mutant lines, dJmj signals were specifically localized to the nucleolus like a wild-type in the late growth stage (). On the other hand, in Pc, Suppressor of zeste12 (Su(z)12), Enhancer of zeste (E(z)) and Lysine(K)-specific demethylase 2 (Kdm2) mutant lines, dJmj signals did not localize at the nucleolus and rather spread throughout the nuclei (). To evaluate more quantitatively whether dJmj signals are localized at the nucleolus, or spread throughout the nuclei, we measured deviation of dJmj signals within spermatocytes nuclei by using Metamorph software. If the fluorescence is concentrated in the nucleolus, signal will be biased in nucleus that gives a low noisiness value (). In contrast, if signals distribute evenly in nucleus, noisiness value will be increased, since the variation of the intensity is low (). By this analysis, we were able to quantitatively evaluate whether dJmj is localized at nucleolus or not. It should be noted that in the Pc (the component of PRC1) mutant line, dJmj signals spread throughout the nucleus ().
Figure 3. Localization of dJmj to the nucleolus in wild-type and PcG mutant lines at late growth stage. (A, C, E, G, I) Phase contrast images of spermatocytes. (B, D, F, H, J) Immunostaining of testes with the anti-dJmj antibody. (A, B) wild-type, (C, D) esc1 : + ; esc1/SM5 ; +, (E, F) ScmET50 : + ; + ; ScmET50/TM3, (G, H) Psc1 : + ; Psc1/CyO ; +, (I, J) ph-d504 : ph-d504/FM7c ; + ; +. (A′–J′) Magnified images of (A-J). (A′–J′) A nucleolus is encircled by dashed line. Scale bar = 50 μm (A-J), 0.1μm (A′–J′).
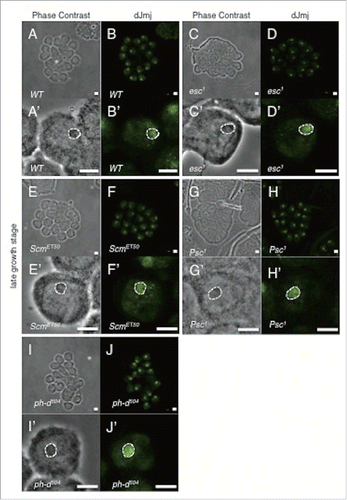
Figure 4. Localization of dJmj to the nucleus in PcG mutant lines. (A, C, E, G) Phase contrast images of spermatocytes at late growth stage. (B, D, F, H) Immunostaining of testes with the anti-dJmj antibody. (A, B) Pc1 : + ; + ; Pc1/TM1, (C, D) Su(z)123 : + ; + ; Su(z)123/TM6B, (E, F) E(z)731 : w ; + ; E(z)731/TM6C, (G, H) E(z)EY21318 : yw ; + ; E(z)EY21318, (I, J) Kdm236Y : w ; + ; Kdm236Y, (A′–J′) Magnified images of (A-J). (A′–H′) A nucleolus is encircled by dashed line. Scale bar = 50 μm (A-H, I, J), 1 μm (I′, J′), 0.1 μm (A′–H′).
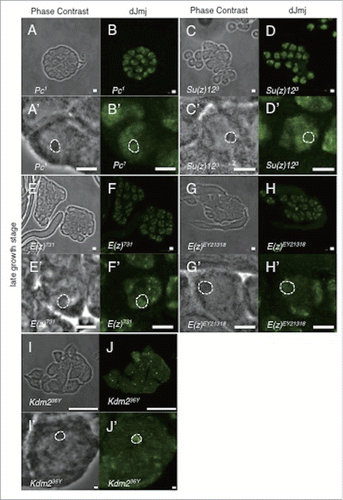
Figure 5. Quantitative analyses of dJmj localization, expression level and nucleolus size in PcG mutant lines at late growth stage. (A) Quantification of dJmj ‘Noisiness’ signals. ‘Noisiness’ is the value which divided average intensity of nuclei by standard deviation of intensity. PcG mutant lines that represent in pink bars were localized to nucleolus. Other mutant lines that represented in blue bar were spread throughout the nucleus. (B) Relative intensity of dJmj signals in the nucleolus. Intensity of dJmj signals in PcG mutant lines was relative to that in wild-type that was scored as 1. (C) Relative size of nucleolus. Average nucleolus size of wild-type was 311.5 μm2. Nucleolus sizes of PcG mutant lines were relative to those of wild-type which were scored as 1. The dJmj signals in PcG mutant lines represented in pink bars were localized to nucleolus. The dJmj signals in other mutant lines represented in blue bars were spread throughout the nucleus. Means of 50 independent measurements on 3 independent preparations are shown. Error bars represent SE. *P < 0.001.
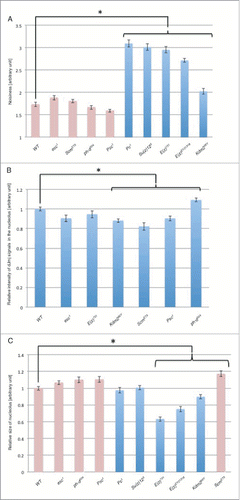
We evaluated not only the localization of dJmj, but also its expression level. To examine the difference of dJmj expression level at the nucleolus in these PcG mutant lines, we quantified relative intensity of dJmj signals in the nucleolus (). In comparison with wild-type, in Scm, Psc, and Kdm2 mutant lines showed significant decrease of the dJmj signal intensity. In contrast, in the ph-d mutant line, dJmj signal in the nucleolus was rather increased.
Nucleolus size difference in the late growth stage in PcG mutant lines
In addition, we measured the size of nucleolus in PcG mutant lines. In E(z) and Kdm2 mutant lines in which dJmj signals spread throughout the nuclei (), nucleolus sizes were smaller than those of the wild-type (). Notably, among these mutant lines, the nucleolus sizes of E(z) mutant line were decreased by more than 35%. These size differences of nucleoli in some of PcG mutant lines are statistically significant.
Localization pattern of dJmj signals at the late growth stage in the tTAF mutant line
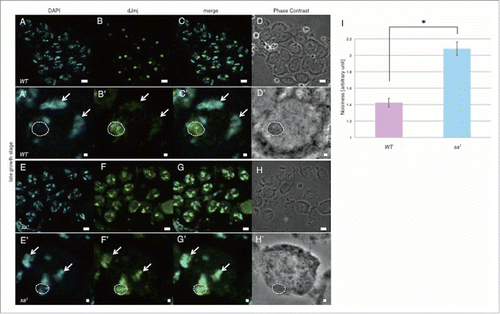
Previous studies indicated that several components of PRC1 such as Pc, Ph and dRing co-localized to the same nucleolar subcompartment with tTAFs in Drosophila spermatocytes, but were not concentrated in the nucleolus in tTAFs mutant spermatocytes.Citation17 Drosophila primary spermatocytes express several tTAFs that control expression of spermatid differentiation genes such as No hitter (Nht, dTAF4L), cannonball (can, dTAF5L), meiosis I arrest (mia, dTAF6L), Spermatocyte arrest (sa, dTAF8L) and Ryan express (Rye, dTAF12L).Citation21 To examine the contribution of tTAFs to dJmj localization at nucleolus in late growth stage, we immunostained testes of the tTAF mutant flies (sa1, bam-GAL4/TM6B) with anti-dJmj antibody. In contrast to nucleolus localization of dJmj in wild-type male testes (), in tTAF mutant flies testes, dJmj signals were observed at both nucleolus and condensed chromatin (). Moreover, we measured deviation of dJmj signals within spermatocyte nuclei by using Metamorph software and confirmed that dJmj significantly localized at not only nucleolus but also chromatin in tTAF mutant line ().
Figure 6. dJmj localization in tTAF mutant line at the late growth stage. (A, E) Staining of spermatocytes with DAPI. (B, F) Immunostaining of testes with the anti-dJmj antibody. (C, G) Merged images are shown. (D, H) Phase contrast images of spermatocytes, (A-D) wild-type male spermatocytes, (E-H) spermatocytes from tTAF mutant line (sa1/TM6B). (A′–H′) Magnified images of each stage. (A′–H′) A nucleus is indicated by arrow and a nucleolus is encircled by dashed line. Scale bar = 10 μm (A-H), 1μm (A′–H′). (I) Quantification of dJmj Noisiness signals in tTAF mutant line. Wild-type that represent in pink bar was localized to nucleolus. tTAF mutant line that represented in blue bar was spread throughout the nucleus. Means of 50 independent measurements on 3 independent preparations are shown. Error bars represent SE. *P < 0.01.
Interaction between Pc and dJmj in the growth stage
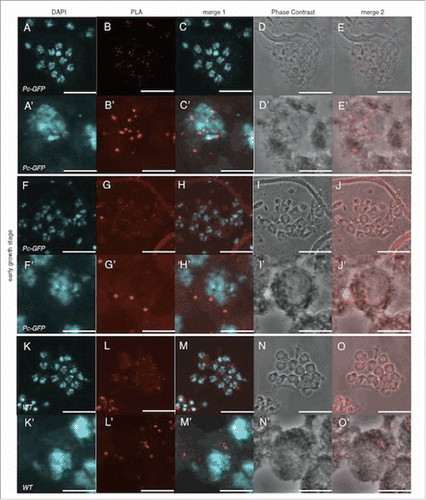
In order to obtain further insight into Pc-dependent nucleolus localization of dJmj, we performed Duolink In Situ Proximity ligation assay (PLA) to examine whether Pc interacts with dJmj at nucleolus in the growth stage. By the Duolink in situ PLA assay, 2 proteins proximate in less than 30 nm can be detectable.Citation22 We used the fly line that expresses Pc-GFP fusion protein as a positive control and wild-type fly as a negative control. In the positive control, using rabbit anti-Pc antibody and mouse anti-GFP antibody as first antibodies, we could detect dotted PLA signals in all nuclei at the early growth stage (). The magnified image of a single cell showed that the intra-molecular Pc and GFP could truly create the signals on the condensed chromatin in this stage (). While in the negative control without expressing Pc-GFP, a few faint signals were detected mainly in cytoplasm even using the same antibodies (). We thus regarded these weak signals as background (). When we used rabbit anti-dJmj antibody and mouse anti-GFP antibody in the fly line that expresses Pc-GFP fusion protein, a similar faint PLA signals were detected in the cytoplasm (). Since Pc localizes on the chromatin and dJmj at periphery of the chromatin in this stage (Fig. S1 A-D′, ), these faint signals in the cytoplasm very likely represent background signals. These results suggest that Pc does not interact with dJmj in the early growth stage.
Figure 7. Pc does not interact with dJmj in the early growth stage. (A-E) Positive control (Pc-GFP line), anti-Pc and anti-GFP antibodies were used for 1st antibodies. (F-J) Sample (Pc-GFP line), anti-dJmj and anti-GFP antibodies were used. (K-O) negative control (wild-type), anti-Pc and anti-GFP antibodies were used. (A, F, K) Staining of spermatocytes with DAPI, (B, G, L) PLA signals, (C, H, M) Merged DAPI staining with PLA signals images are shown. (E, J, O) Merged PLA signals with Phase contrast images are shown. (D, I, N) Phase contrast images of spermatocytes, (A′–O′) Magnified images of (A-O), Scale bar = 10 μm.
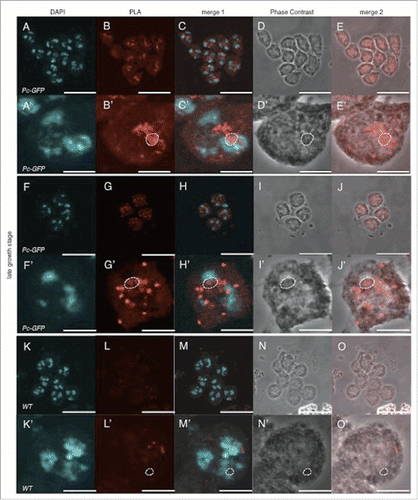
In the late growth stage, both Pc and dJmj signals were exclusively concentrated at the nucleolus with weak signals throughout nucleus (Fig. S1 E-H′, ). In the positive control with both anti-Pc and anti-GFP antibodies, PLA signals were mainly detected in both nucleolus and nucleoplasm with a few in cytoplasm (). While in negative control, a few faint signals were detected in nucleoplasm (). When we used both anti-dJmj and anti-GFP antibodies in the fly expressing Pc-GFP, PLA signals were detected in both nucleolus and nucleoplasm with a few in cytoplasm (). These results suggest that Pc interacts with dJmj in both nucleolus and nucleoplasm at the late growth stage.
Figure 8. Signals that represent the interaction between Pc and dJmj in the late growth stage. (A-E) Positive control (Pc-GFP line), anti-Pc and anti-GFP antibodies were used for 1st antibodies. (F-J) Sample (Pc-GFP line), anti-dJmj and anti-GFP antibodies were used. (K-O) negative control (wild-type), anti-Pc and anti-GFP antibodies were used. (A, F, K) Staining of spermatocytes with DAPI, (B, G, L) PLA signals, (C, H, M) Merged DAPI staining with PLA signals images are shown. (E, J, O) Merged PLA signals with Phase contrast images are shown. (D, I, N) Phase contrast images of spermatocytes, (A′–O′) Magnified images of (A-O). (B′–E′, G′–J′, L′–O′) A nucleolus is encircled by dashed line. Scale bar = 10 μm.
Subcellular localization of H3K27me3 in testes during spermatogenesis and spermiogenesis
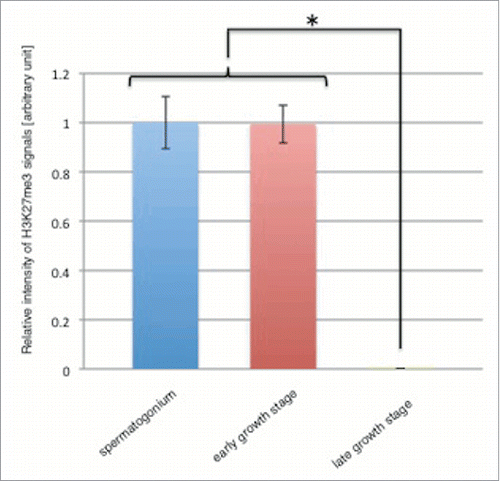
H3K27me3 is a well-known marker for PcG dependent repressive state of chromatin.Citation23 To detect H3K27me3 during spermatogenesis and spermiogenesis, we immunostained wild-type male testes with monoclonal antibody to H3K27me3.Citation24 In the spermatogonium and the early growth stage, H3K27me3 signals were exclusively localized in nucleus with no signal in the cytoplasm (). At the following late growth stage, H3K27me3 signals were detected in all of chromatin clumps with no signal in nucleolus (). No such H3K27me3 signal was detectable in the control experiments with the second antibody only (). In the next onion stage, H3K27me3 signals were distributed in the nuclei with no signal in cytoplasm (). Unlike dJmj signals, H3K27me3 signals were detected in the nuclei at the elongation stage (). In the canoe stage, H3K27me3 signal was no more detectable in both cytoplasm and nucleus (). Interestingly, quantitative analyses revealed that the level of H3K27me3 in whole nuclei extensively decreased at the late growth stage ().
Figure 9. H3K27me3 level decreases in the late growth stage. Relative intensity of H3K27me3 signals in spermatogonium (blue bar), early growth stage (red bar) and late growth stage (green bar) are shown. Intensity of H3K27me3 signals in the growth stage was relative to that in spermatogonium that was scored as 1. Means of 49 independent measurements on 3 independent preparations are shown. Error bars represent SE. *P < 0.001.
Discussion
The Drosophila genome contains at least 13 genes encoding JmjC domain-containing proteins.Citation25 However, little is known about their biological roles and their contributions to chromatin regulation. In the present study, we showed that subcellular localization of dJmj changes dramatically during spermatogenesis. In spermatogonium, dJmj localizes in both nucleus and perinuclear region of the cytoplasm (). In the early growth stage of spermatocyte, dJmj signals characteristically appeared as dotted signals at periphery of the chromatin (), although the biological meaning of this appearance is not known yet. These results suggest that somehow dJmj may be functional in chromatin regulation at these stages.
In the following late growth stage, dJmj is transported into nucleus, but the signal mainly localizes at nucleolus (). The nucleolus localization of dJmj depends on PcG genes such as Pc, Su(z)12, E(z) and Kdm2 (). Interestingly, previous reports showed that Pc and other components of PRC1 are co-localized to the same nucleolar subcompartment with tTAFs in spermatocytes.Citation17 Furthermore, localization of PRC1 components to the spermatocyte nucleolus is coincident with tTAFs expression and dependent on wild-type tTAFs function.Citation17 These results suggest that tTAFs act as derepressors by sequestering PRC1 to the spermatocyte nucleolus to counteract PcG-induced repression. However, other studies indicate that Pc and tTAFs associate with chromosome masses and also accumulate in clumps at the periphery of the nucleolus or coat the nucleolar surface appearing as a ring around the nucleolus in the spermatocytes.Citation18 In the present study, we detected Pc mainly in nucleolus at late growth stage (Fig. S1 E-H′). Corresponding to it, PLA signals between Pc and dJmj were detected in both nucleolus and nucleoplasm with a few in cytoplasm suggesting that dJmj interacts with Pc in nucleolus at the late growth stage ().
In the following onion stage, dJmj signals were distributed in both nucleus and perinuclear region of the cytoplasm (). Since gene expression is extremely low in onion stage, dJmj may play a role in chromatin regulation at this stage that is not directly related to gene expression. In the elongation and canoe stages, dJmj signal became no more detectable in both cytoplasm and nuclei ().
H3K27me3 signals were exclusively detectable in nuclei but not in nucleoli throughout spermatogenesis (). However, quantitative analyses revealed that the level of H3K27me3 in whole nuclei extensively decreased at the late growth stage (). In the growth stage, many genes are activated and expressed in the nucleus to support rapid growth of spermatocyte.Citation21 Since dJmj that is supposed to be a negative regulator of gene expression is not required in this stage, it might be recruited into nucleolus in cooperation with these factors. Therefore, compartmentalization of dJmj in nucleolus together with some of PcG could decrease H3K27me3 level in nuclei that likely results in de-repression of the expression of genes required to cellular growth and proliferation in the following meiotic divisions.
dJmj localization pattern in the late growth stage of spermatocyte in PcG mutant lines could be divided in 3 groups. In the first group, dJmj localized at the nucleolus similarly to the wild-type spermatocyte (). Therefore PcG genes that belong to this group, esc, Scm, Psc, and ph-d may not play a role in dJmj recruitment to nucleolus ().
In the second group, dJmj signal localized at the whole nuclei (). The genes belong to this group such as Pc, Su(z)12, E(z) and Kdm2 genes play a role in dJmj recruitment to nucleolus. Interestingly in some of these PcG mutant lines (E(z) and Kdm2 mutant lines), nucleolus sizes were smaller than those of wild-type (). Notably, among these factors, the nucleolus sizes of E(z) mutant line was decreased by more than 35%. These results implicate that these factors together with dJmj are involved in nucleolus growth. However, since the effect on nucleolus growth could be a secondary effect, these factors may not be directly involved in regulation of nucleolus growth. Further analyses are necessary to clarify this point.
In the third group, the intensity of dJmj signals was different from that of wild-type (). Scm, Psc and Kdm2 mutant lines showed significant decrease of the dJmj signal intensity in comparison with that of wild-type. However, in the ph-d mutant line, dJmj signal was rather increased. Therefore the ph-d gene may act as negative regulator of dJmj expression. Taken together, the factors that belong to this group are required to fine-tune global dJmj level.
Up to now, the mechanism for nucleolus localization of proteins is poorly understood. Strikingly specific localization of dJmj in nucleolus at late growth stage of spermatogenesis will provide an useful system to study detailed mechanism for nucleolus localization.
Materials and methods
Fly stocks
Fly stocks were cultured at 25°C on standard food. The Canton S fly was used as the wild-type strain. Pc-GFP fly stock (w; Pc-GFP) was obtained from Dr. Renato Paro. Spermatocyte arrest (sa) mutant fly stock (sa1) and all other stocks used in this study were obtained from the Bloomington Drosophila stock center, Drosophila Genetic Resource Center in Kyoto, and Exelixis Collection. The sa1, bam-GAL4/TM6B flies are described previously.Citation26 The details of PcG mutant alleles used in immunostaining analyses are described below; esc: esc1 (hypomorphic allele). Scm: ScmET50 (hypomorphic allele). Psc: Psc1 (hypomorphic allele). Ph-d: ph-d504 (loss of function). Pc: Pc1 (amorphic allele). Su(z): Su(z)123 (point mutation by ethyl methanesulfonate (EMS) on 3L ; 19,921,738). E(z): E(z)731 (point mutation by EMS on 3L; 10,635,007), E(z)EY21318 (P-element insertion on 3L; 10,637,896). Kdm2: Kdm236Y (P-element insertion on first intron of Kdm2).
Testis sample preparation
Testis sample preparation was carried out according to the method of Inoue et al.Citation27 We dissected testes in Testis Buffer (TB) (10mM Tris-Cl, pH6.8, 183mM KCL, 47mM NaCl, 1mM EDTA) from young adults. We transferred the testes in a drop of TB on a coverslip and tore them open using tungsten needles. Then, we place a slide over the coverslip without any pressing. It made testis cells flatten and affixed them on to the slide. The testis samples were provided for immunostaining experiments after methanol fixation.
Immunostaining of testis
The testis samples were placed in liquid nitrogen, then were fixed in methanol for 5 min at −20°C and in acetone for 5 min at −30°C and washed with PBST (0.1% TritonX-100 in PBS) for 10 min, 3 times in PBS for 5 min. The samples were blocked in 0.15% TritonX-100 in PBS containing 10% goat serum at 25°C for 30 min and incubated with primary antibodies for 16 hours at 4°C. The samples were then incubated with secondary antibodies for 2.5 hours at 25°C. The primary antibodies used in this study were anti-dJmj (1:200),Citation19 H3K27me3 (1:500),Citation24 anti-GFP (1:150) (Nacalai Tesque), anti-Fibrillarin (1:200) (Abcam). Secondary antibodies were Alexa Fluor 488 anti-rabbit IgG (1:400), Alexa Flour 546 anti-mouse IgG (1:400) (Molecular Probes). The samples were washed 3 times in PBS for 5 min and once in H2O for 1 min. DNA was stained with DAPI. Preparations were examined using a confocal laser scanning microscope (Olympus FV10i) and analyzed by using Metamorph software (Molecular Devices).
Noisiness analysis
Region measurements function of Metamorph software (Molecular Devices) was used for this analysis. Noisiness analysis indicates intensity signal/noise, a measure of the inherent “noisiness” of the region or entire image, calculated by dividing the average intensity value by the standard deviation. This analysis is suitable to evaluate specific localization of signals in cells, even though intensity of signals in each cell varies to some extent.
Duolink In Situ proximity ligation assay (PLA)
Duolink In situ PLA kits were purchased from Sigma-Aldrich. Fixation of testes, blocking of nonspecific binding of antibody and treatment with anti-GFP, anti-dJmj or anti-Pc antibodies were done as described above. Subsequently, secondary antibodies conjugated with the PLA oligonucleotide probes were used. Duolink PLA probes (Duolink In Situ PLA probe anti-mouse PLUS and anti-rabbit MINUS antibody, Antibody Diluent (Sigma-Aldrich)) were pre-incubated at 25°C for 40 min and then incubated at 37°C for 1 hour. Ligation of the connector oligonucleotides were done with Ligation Solution including Ligase (1 unit/uL) (Sigma-Aldrich) at 37°C for 30 min with shaking. Rolling-circle amplification and detection of the amplified DNA products were done with Amplification Solution including DNA Polymerase (10 unit/uL) (Sigma-Aldrich) at 37°C in dark for 100 min with shaking. DNA was stained with DAPI. Preparations were examined using a confocal laser scanning microscopy (Olympus FV10i).
Statistical analysis
Unpaired two-tailed Student t-tests were performed to analyze the results of immunostaining analyses. Compared results were considered statistically significant when the P-value < 0.01.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KSPE_S_1232023.zip
Download Zip (737.9 KB)Funding
This study was partially supported by Dr. Renato Paro, Department of Biosystems Science and Engineering, ETH Zürich, Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, JST and the JSPS Core-to-Core Program, Asia-Africa Science Platforms, JSPS KAKENHI Grant Number 15J0948.
ORCID
Hiroshi Kimura http://orcid.org/0000-0003-0854-083X
References
- Balciunas D, Ronne H. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem Sci 2000; 25:274-6; PMID:10838566; http://dx.doi.org/10.1016/S0968-0004(00)01593-0
- Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci 2001; 26:7-9; PMID:11165500; http://dx.doi.org/10.1016/S0968-0004(00)01700-X
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev 1995; 9:1211-22; PMID:7758946; http://dx.doi.org/10.1101/gad.9.10.1211
- Ahmed S, Palermo C, Wan S, Walworth NC. A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast. Mol Cell Biol 2004; 24:3660-9; PMID:15082762; http://dx.doi.org/10.1128/MCB.24.9.3660-3669.2004
- Ayoub N, Noma K, Isaac S, Kahan T, Grewal SIS, Cohen A. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol 2003; 23:4356-70; PMID:12773576; http://dx.doi.org/10.1128/MCB.23.12.4356-4370.2003
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 2005; 18:623-35; PMID:15949438; http://dx.doi.org/10.1016/j.molcel.2005.05.012
- Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn 2005; 232:21-32; PMID:15580614; http://dx.doi.org/10.1002/dvdy.20204
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn 2006; 235:2449-59; PMID:16715513; http://dx.doi.org/10.1002/dvdy.20851
- Herz H-M, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, et al. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol Cell Biol 2012; 32:1683-93; PMID:22354997; http://dx.doi.org/10.1128/MCB.06503-11
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009; 461:762-7; PMID:19767730; http://dx.doi.org/10.1038/nature08398
- Klymenko T, Müller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 2004; 5:373-7; PMID:15031712; http://dx.doi.org/10.1038/sj.embor.7400111
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 2007; 8:9-22; PMID:17173055; http://dx.doi.org/10.1038/nrg1981
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 2010; 24:368-80; PMID:20123894; http://dx.doi.org/10.1101/gad.1886410
- Cheng J, Türkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature 2008; 456:599-604; PMID:18923395; http://dx.doi.org/10.1038/nature07386
- Dorogova NV, Akhmametyeva EM, Kopyl SA, Gubanova NV, Yudina OS, Omelyanchuk LV, Chang L-S. The role of Drosophila Merlin in spermatogenesis. BMC Cell Biol 2008; 9:1; PMID:18186933; http://dx.doi.org/10.1186/1471-2121-9-1
- Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci 2007; 120:1689-700; PMID:17452629; http://dx.doi.org/10.1242/jcs.004663
- Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 2005; 310:869-72; PMID:16272126; http://dx.doi.org/10.1126/science.1118101
- El-Sharnouby S, Redhouse J, White RAH. Genome-wide and cell-specific epigenetic analysis challenges the role of polycomb in Drosophila spermatogenesis. PLoS Genet 2013; 9:e1003842; PMID:24146626; http://dx.doi.org/10.1371/journal.pgen.1003842
- Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M. The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J 2007; 274:6139-51; PMID:17970746; http://dx.doi.org/10.1111/j.1742-4658.2007.06135.x
- Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 2008; 454:241-5; PMID:18528333; http://dx.doi.org/10.1038/nature07014
- Hiller M, Xin C, Jodeane P, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, et al. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 2004; 131:5297-308; PMID:15456720; http://dx.doi.org/10.1242/dev.01314
- Kitazawa D, Matsuo T, Kaizuka K, Miyauchi C, Hayashi D, Inoue YH. Orbit/CLASP is required for myosin accumulation at the cleavage furrow in Drosophila male meiosis. PLoS ONE 2014; 9:e93669; PMID:24850412; http://dx.doi.org/10.1371/journal.pone.0093669
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49:808-24; PMID:23473600; http://dx.doi.org/10.1016/j.molcel.2013.02.013
- Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct 2008; 33:61-73; PMID:18227620; http://dx.doi.org/10.1247/csf.07035
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7:715-27; PMID:16983801; http://dx.doi.org/10.1038/nrg1945
- Kitazawa D, Yamaguchi M, Mori H, Inoue YH. COPI-mediated membrane trafficking is required for cytokinesis in Drosophila male meiotic divisions. J Cell Sci 2012; 125:3649-60; PMID:22553212; http://dx.doi.org/10.1242/jcs.103317
- Inoue YH, Savoian MS, Suzuki T, Mathe E, Yamamoto M, Glover DM. Mutations in orbit/mast reveal the central spindle is comprised for two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J Cell Biol 2004; 166:49-60; PMID:15240569; http://dx.doi.org/10.1083/jcb.200402052

