ABSTRACT
Cell biology is increasingly evolving to become a more formal and quantitative science. The field of intracellular transport is no exception. However, it is extremely challenging to formulate mathematical and computational models for processes that involve dynamic structures that continuously change their shape, position and composition, leading to information transfer and functional outcomes. The two major strategies employed to represent intracellular trafficking are based on “ordinary differential equations” and “agent-” based modeling. Both approaches have advantages and drawbacks. Combinations of both modeling strategies have promising characteristics to generate meaningful simulations for intracellular transport and allow the formulation of new hypotheses and provide new insights. In the near future, cell biologists will encounter and hopefully overcome the challenge of translating descriptive cartoon representations of biological systems into mathematical network models.
Moving towards a more formal and quantitative cell biology
Cells are extraordinarily complex nano-machines capable of performing a broad set of functions, each subject to sophisticated regulatory control. Despite the exponential growth in our knowledge of the molecular mechanisms underlying cell functions, our understanding of how these machines work is in many cases fragmentary and qualitative. Our hypotheses about the function of a given factor in a cell process are usually expressed as “it is involved“, “it is required“, “it participates“, “it is part of the mechanism“, and so on. Only seldom the function is described in detail, quantitatively, systematically and inserted in a well-defined, formally described cellular mechanism. In other words, following the intuitive definition of models proposed by GunawardenaCitation1 (), most cell biologists build their research and design their experiments based on informal (or conceptualCitation2) models.
Table 1. Gunawardena definitions for formal and informal model.
Moving cell biology to a more quantitative discipline is, in essence, progressing our thinking and methodologies beyond the purely empirical to incorporate mathematical models in our research. This necessity has been emphasized numerous times.Citation3–5 Remarkable efforts in moving cell biology in this new direction are exemplified by excellent books – Cell Biology by the NumbersCitation6 and Physical Biology of the CellCitation7 and a slew of recent papers (ex.Citation8). Even approximated estimations of absolute number of molecules, cellular structures and organelles, kinetic and dynamic parameters, will be fundamental for a thorough understanding of the mechanisms underlying the sophisticated behavior of the cell.
Computational cell biology is an advancing field, where different mathematical tools are employed to analyze and organize the large amount of data generated with advanced technologies such as imaging, omics and high throughput screening. Hence, an increasingly large group of scientists trained in computer science, mathematics, statistics and data science are now entering the field of cell biology. This is a remarkable opportunity to advance cell biology towards a more formal science.
Developing formal models for cellular processes is at the interface of cell biology and physics/math/computer science. An appropriate selection of mathematical tools, along with the specification of the biological hypothesis is required to build these models. Some fundamental questions that need to be addressed include, but are not limited to: (1) what kind of model to use; (2) what elements should be included; (3) what is the functional interplay between the elements; and (4) what set of parameters is to be included. Thus, the development of formal models for cellular processes opens up a promising field for synergistic interaction between the experimentalists and the physics/math/computer science-oriented scientists. Such interactions between diverse groups of scientists with their unique skill sets, will foster the development of a new, more formal and quantitative field of cell biology.
Formal models for intracellular transport
Computational modeling tools are essential for understanding the underlying mechanisms and emergent properties in cell biology, and the field of intracellular trafficking is no exception. However, the dynamic nature of organelles makes the formulation of hypotheses challenging since it is difficult to develop simple models for the organelles of the endocytic and secretory pathways: these organelles change position, shape, and undergo processes such as fusion and fission in addition to simultaneously altering their composition through a complex network of interactions among lipids and membrane-associated and soluble proteins.
Available methods for model development
Currently, a large set of computational modeling technologies and tools are available,Citation9 but not all are user-friendly and most require at least some basic programming skills. In regard to modeling based on Differential Equations (DE), few exceptions exist that include powerful and easy-to-use platforms for computational cell biology. For example, COPASICitation10 provides user friendly interfaces for ordinary differential equations (ODE)s, and Cell DesignerCitation11 allows the design of complex interacting networks of factors using simple and intuitive graphical notations. Both platforms are fully compliant with the Systems Biology Markup Language (SBML),Citation12 a standard for expressing mathematical models. Therefore, COPASI and Cell Designer can be used in combination with other tools compatible with this language.
A limiting factor for most of the DE modeling platforms is that the network of interactions is examined in a homogeneous space. However, some software can be used to simulate the spatio–temporal dynamics of processes in a cell, e.g., Virtual Cell (VCell).Citation13 In VCell, the shape of the compartments can be specified by analytical geometry equations, or can be derived from imported images, such as 3D confocal microscope stacks. Then, the spatial and temporal behaviors of substances can be analyzed in a relatively easy to use program. However, compartments in VCell are static structures, and what is recorded is the diffusion and molecular interactions occurring at different coordinates of the model. This is a significant limitation for intracellular transport where the position and shape of organelles change continuously as the membrane-bound structures move in the cytosol and undergo fusion with other organelles and fission by pinching off vesicles and tubules.
Another modeling technology, agent-based modeling (ABM) uses “agents” to represent the elements in biological processes. An agent can be anything from a single molecule, such as a receptor or a ligand, to a complete organelle. Agents can have different composition, properties, undergo dynamic changes, including movements in 2D or 3D spaces. The change in the agents or the interactions between agents can be specified with simple rules. For example:
“If there is an endosome in a given position, and another endosome is present nearby, and if their membrane domains are compatible, fuse them to form a single organelle containing all the membrane-associated and soluble components of the original two endosomes”.
A valuable characteristic of the ABM methods is that the rules are somehow intuitive and approximate the way of thinking in informal models. Another advantage is that the “visualization” of the process is meaningful for cell biologists (). Notwithstanding, the huge flexibility of the ABM models has its drawbacks. Agents can be almost anything; hence the platforms for modeling lack specificity and in order to model a cell biological process, the rules need to be programmed. This in turn requires more than basic knowledge of programming. There are numerous ABM softwares with different characteristics that are freely available (https://en.wikipedia.org/wiki/Comparison_of_agent-based_modeling_software). NetLogo has an especially simple coding language; yet, basic programming skills are necessary (https://ccl.northwestern.edu/netlogo/). Repast is a more flexible and general platform, but it requires the working knowledge of Java or C++ (https://sourceforge.net/p/repast/wiki/Home/). In contrast to DE methods, ABMs lack a common language with common syntaxes that would make the rules, and hence the underlying hypotheses, transparent. Finally, although molecular interactions and chemical reactions can be programmed using “rules” in ABM, this is not accomplished as easily as with DE-based models, especially when complex networks of interactions are to be modeled.
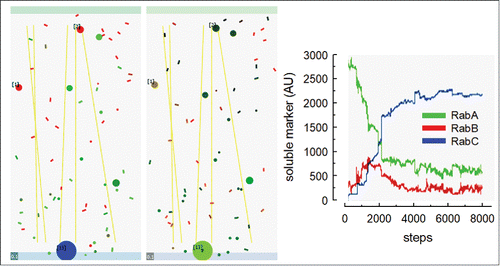
Figure 1. ABM simulation of intracellular transport modeled in NetLogo. The ABM “rules” for fusion and fission are described in.Citation28 In brief, three endocytic compartments carrying different Rab membrane domains were modeled. Each compartment is formed by several individual organelles (tubules and vesicles, represented by rectangles and circles in the right and middle panels). The organelles move randomly (or along microtubules, yellow straight lines) in the 2D space. Two nearby organelles can fuse if they carry compatible domains. An organelle can divide forming a tubule and a vesicle, or generate an internal vesicle if it contains enough membrane for these topological transformation. Rabs membrane domains change composition by series of reactions involving cytosolic Rabs. At step 0, a soluble and a membrane bound markers were loaded in the Rab A compartment. Left panel. Snapshot after 8380 steps. In each step, organelles have the opportunity of moving, fusing, dividing, forming an internal vesicles, and changing Rab composition. The color of each organelle (tubule or vesicle) represents its Rab content: Rab A (green), Rab B (red) and Rab C (blue). The resulting color depends on the relative amount of these proteins on the membranes. For example, the large organelle at the bottom is enriched in Rab C. Middle panel. Same simulation at step 8385. Color code was changed to reflect the content of the fluid phase (green) or the membrane-associated (red) markers. Notice that the soluble marker is concentrated in the large organelle labeled with Rab C and that the membrane-associated marker is limited to small tubules. Black organelles lack soluble or membrane markers. Plasma membrane is at the top and nucleus at the bottom. The width of the gray square at the left bottom correspond to 100 nm. Numbers in brackets indicate the amount of internal vesicles of each organelle. Right panel. The graph shows the distribution of the fluid phase marker (in arbitrary units) associated with Rab A (green), Rab B (red) and Rab C (blue) organelles along the simulation. Notice that the marker is initially in Rab A structures; along the simulation it is transferred to other organelles and finally concentrates in Rab C structures.
The combination of ABM and DE-based models is not frequent.Citation14,15 The ENteric Immunity Simulator (ENISI)Citation16 is a simulator of the gastrointestinal tract mucosal immune responses, where cells are modeled as agents in Repast and complex signal transduction networks are solved in COPASI. The combination of the two methods has proved to be a very powerful tool to identify key cells and molecular factors that modulate the immune response to H. pyloriCitation17,18 and C. difficile.Citation19
Examples of published models for intracellular transport
Most papers published about the molecular mechanisms of intracellular transport do not include an explicit mathematical model. In general, the informal models proposed to represent the findings are limited to select relevant components distributed in a few organelles and arrows indicating either transport or interactions. Although comparatively scarce, several formal models for intracellular transport have been published, in most cases, as proof of principle of different transport mechanisms. This is particularly evident for the formal models addressing the maturation/vesicular transport controversy surrounding Golgi biogenesis and transport.Citation20-22 Formal models have also been used to distinguish between different hypotheses compatible with experimental data.Citation23,24
Most of the published models to describe intracellular transport use ODE. A seminal example is the paper by Heinrich and RapoportCitation25 where they showed that transport among stable non-identical compartments can be achieved by a combination of SNARE-dependent fusion and coat-dependent budding. Less abundant are models based on ABM. The spatial and temporal dynamics of autophagic organelles, and the cellular network of mitochondria have been simulated using ABM implemented in NetLogo.Citation26,27 In addition, the efficiency and basic requirement of fusion/fission-dependent intracellular transport of fluid phase markers was assessed by an ABM programmed in Visual Basics.Citation28 More mechanistic ABMs have been used to analyze the molecular crowding effect and the influence of the cytoskeleton on intracellular transport.Citation29,30 Integrated models that combine ABM and DE are rare; an example is the development of a model to study mitophagy,Citation31 in MATLAB.
Concluding remarks
Tools for modeling using ODE are freely available and are relatively user-friendly. Networks of molecular interactions, or signaling cascades can be easily modeled without significant formal training in computer programming. Platforms for ABM, on the other hand, are too generic to be used to generate formal models for intracellular transport, and would need some basic programming skills. Thus, there is an urgent need to develop more user-friendly tools in the future to allow even the modeling novice to integrate mathematical approaches into their research. However, the emergence of computational tools alone will not be sufficient to develop and formalize the problems in quantitative cell biology. It is crucial that the researchers start to adapt and transform their conceptual way of thinking about the biological processes into more formal mathematical models. In order to do that, training modules and workshops are necessary to facilitate interaction between cell biologists and modeling/mathematical scientists (ex: the recently organized “Finding your inner modeler” workshop described in this volume of Cellular Logistics). In essence, building a formal model is not harder than to propose a thoughtful informal one. Initially, the parameter space that needs to be estimated may seem daunting to an experimentalist, but soon he/she will realize that most of them can be narrowed to an acceptable range, based on bibliography or common sense, or can be derived from “doable” experiments. Another concern is that there may be too many unknowns in the process. In the informal models, these undisclosed parts of the mechanism are treated as a black box (we have some understanding of its behavior, but not of its composition or mechanistic details). Thankfully, the same strategy can be programmed into a formal model. Finally, formal models are easier to be falsified than informal ones. This is an extremely valuable characteristic, but only if we are ready to update/modify/discard our models when they are inconsistent with experimental data. As written by Gunawardena “…formal models are not descriptions of reality; they are descriptions of our assumptions about reality; they are only as good as their assumptions…”.Citation1
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We greatly appreciate the critical comments of Claudia Tomes and Ignacio Cebrián.
Funding
We thank the support of a Fulbright fellowship granted by the Ministerio de Educación y Deportes, Argentina and a Beca de Movilidad Docente and the grant 06/J478 from the Universidad Nacional de Cuyo, Argentina.
References
- Gunawardena J. Beware the tail that wags the dog: informal and formal models in biology. Mol Biol Cell. 2014;25:3441–4. doi:10.1091/mbc.E14-02-0717. PMID:25368417.
- Howard J. Quantitative cell biology: the essential role of theory. Mol Biol Cell. 2014;25:3438–40 doi:10.1091/mbc.E14-02-0715. PMID:25368416.
- Mogilner A, Wollman R, Marshall WF. Quantitative modeling in cell biology: what is it good for? Dev Cell. 2006;11:279–87. doi:10.1016/j.devcel.2006.08.004. PMID:16950120.
- Lippincott-Schwartz J. Quantitative cell biology: transforming the conceptual, theoretical, instrumental, and methodological approaches to cell biology. Mol Biol Cell. 2014;25:3437–1297. doi:10.1091/mbc.E14-08-1297. PMID:25368415.
- Flamholz A, Phillips R, Milo R. The quantified cell. Mol Biol Cell. 2014;25:3497–500. doi:10.1091/mbc.E14-09-1347. PMID:25368429.
- Milo R, Phillips R. Cell biology by the numbers. New York: Garland Science; 2016.
- Phillips R, Kondev J, Theriot J, Orme N. Physical biology of the cell. 2nd ed. New York: Garland Science; 2013.
- Vodovotz Y, Xia A, Read EL, Bassaganya-Riera J, Hafler DA, Sontag E, Wang J, Tsang JS, Day JD, Kleinstein SH, et al. Solving Immunology? Trends Immunol. 2017;38:116–27 doi:10.1016/j.it.2016.11.006. PMID:27986392.
- Bassaganya-Riera J. Computational Immunology: Models and Tools. Amsterdam: Elsevier, Academic Press; 2016.
- Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI–a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–74. doi:10.1093/bioinformatics/btl485. PMID:17032683.
- Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol. 2005;23:961–6. doi:10.1038/nbt1111. PMID:16082367.
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi:10.1093/bioinformatics/btg015. PMID:12611808.
- Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19:401–6. doi:10.1016/S0167-7799(01)01740-1. PMID:11587765.
- Mei Y, Carbo A, Hoops S, Hontecillas R, Bassaganya-Riera J. ENISI SDE: A New Web-Based Tool for Modeling Stochastic Processes. IEEE/ACM Trans Comput Biol Bioinform. 2015;12:289–97. doi:10.1109/TCBB.2014.2351823. PMID:26357217.
- Sutterlin T, Kolb C, Dickhaus H, Jager D, Grabe N. Bridging the scales: semantic integration of quantitative SBML in graphical multi-cellular models and simulations with EPISIM and COPASI. Bioinformatics. 2013;29:223–9 doi:10.1093/bioinformatics/bts659. PMID:23162085.
- Mei Y, Carbo A, Hontecillas R, Hoops S, Liles N, Lu P, Philipson C, Bassaganya-Riera J. ENISI MSM: A novel multi-scale modeling platform for computational immunologyBioinformatics and Biomedicine (BIBM). IEEE International Conference. IEEE; 2014 p. 391–6
- Leber A, Hontecillas R, Tubau-Juni N, Zoccoli-Rodriguez V, Hulver M, McMillan R, Eden K, Allen IC, Bassaganya-Riera J. NLRX1 Regulates Effector and Metabolic Functions of CD4+ T Cells. J Immunol. 2017;198:2260–68 doi:10.4049/jimmunol.1601547. PMID:28159898.
- Leber A, Bassaganya-Riera J, Tubau-Juni N, Zoccoli-Rodriguez V, Viladomiu M, Abedi V, Lu P, Hontecillas R. Modeling the role of lanthionine synthetase C-Like 2 (LANCL2) in the modulation of immune responses to helicobacter pylori infection. PLoS ONE. 2016;11:e0167440 doi:10.1371/journal.pone.0167440. PMID:27936058.
- Leber A, Viladomiu M, Hontecillas R, Abedi V, Philipson C, Hoops S, Howard B, Bassaganya-Riera J. Systems modeling of interactions between mucosal immunity and the gut microbiome during clostridium difficile infection. PLoS ONE. 2015;10:e0134849 doi:10.1371/journal.pone.0134849. PMID:26230099.
- Patterson GH, Hirschberg K, Polishchuk RS, Gerlich D, Phair RD, Lippincott-Schwartz J. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell. 2008;133:1055–67. doi:10.1016/j.cell.2008.04.044. PMID:18555781.
- Sachdeva H, Barma M, Rao M. Nonequilibrium description of de novo biogenesis and transport through Golgi-like cisternae. Sci Rep. 2016; 19;6:38840. doi:10.1038/srep38840. PMID:27991496.
- Mani S, Thattai M. Stacking the odds for Golgi cisternal maturation. Elife. 2016;5:e16231. doi:10.7554/eLife.16231. PMID:27542195.
- Jordao L, Bleck CK, Mayorga L, Griffiths G, Anes E. On the killing of mycobacteria by macrophages. Cell Microbiol. 2008;10:529–48 PMID:17986264.
- Kalaidzidis I, Miaczynska M, Brewinska-Olchowik M, Hupalowska A, Ferguson C, Parton RG, Kalaidzidis Y, Zerial M. APPL endosomes are not obligatory endocytic intermediates but act as stable cargo-sorting compartments. J Cell Biol. 2015;211:123–44. doi:10.1083/jcb.201311117. PMID:26459602.
- Heinrich R, Rapoport TA. Generation of nonidentical compartments in vesicular transport systems. J Cell Biol. 2005;168:271–80. doi:10.1083/jcb.200409087. PMID:15657397.
- Borlin CS, Lang V, Hamacher-Brady A, Brady NR. Agent-based modeling of autophagy reveals emergent regulatory behavior of spatio-temporal autophagy dynamics. Cell Commun Signal. 2014;12:56. doi:10.1186/s12964-014-0056-8. PMID:25214434.
- Dalmasso G, Marin Zapata PA, Brady NR, Hamacher-Brady A. Agent-Based Modeling of Mitochondria Links Sub-Cellular Dynamics to Cellular Homeostasis and Heterogeneity. PLoS ONE. 2017;12:e0168198 doi:10.1371/journal.pone.0168198. PMID:28060865.
- Mayorga LS, Campoy EM. Modeling fusion/fission-dependent intracellular transport of fluid phase markers. Traffic. 2010;11:1001–15. doi:10.1111/j.1600-0854.2010.01067.x. PMID:20374555.
- Klann M, Koeppl H, Reuss M. Spatial modeling of vesicle transport and the cytoskeleton: the challenge of hitting the right road. PLoS ONE. 2012;7:e29645 doi:10.1371/journal.pone.0029645. PMID:22253752.
- Klann MT, Lapin A, Reuss M. Agent-based simulation of reactions in the crowded and structured intracellular environment: Influence of mobility and location of the reactants. BMC Syst Biol. 2011;5:71. doi:10.1186/1752-0509-5-71. PMID:21569565.
- Choe SC, Hamacher-Brady A, Brady NR. Autophagy capacity and sub-mitochondrial heterogeneity shape Bnip3-induced mitophagy regulation of apoptosis. Cell Commun Signal. 2015;13:37. doi:10.1186/s12964-015-0115-9. PMID:26253153.

