ABSTRACT
The current pandemic called COVID-19 caused by the SARS-CoV-2 virus brought the need for the search for fast alternatives to both control and fight the SARS-CoV-2 infection. Therefore, a race for a vaccine against COVID-19 took place, and some vaccines have been approved for emergency use in several countries in a record time. Ongoing prophylactic research has sought faster, safer, and precise alternatives by redirecting knowledge of other vaccines, and/or the development of new strategies using available tools, mainly in the areas of genomics and bioinformatics. The current review highlights the development of synthetic antigen vaccines, focusing on the usage of bioinformatics tools for the selection and construction of antigens on the different vaccine constructions under development, as well as strategies to optimize vaccines for COVID-19.
The current pandemic called COVID-19 caused by the SARS-CoV-2 virus is responsible for over 200 million cases and 4 million deaths.Citation1 It has also brought the need for new political, economic, and social perspectives which maximize the search for fast alternatives to both control and fight the SARS-CoV-2 infection. Therefore, a race for a vaccine against COVID-19 took place, and in less than a year, some of the studies have reached phase 3 of vaccine trials as well as some others have been approved for emergency use in several countries.Citation2,Citation3 Ongoing prophylactic research has sought faster, safer, and precise alternatives that can be reached by redirecting knowledge of other vaccines that already exist for other diseases, and/or the development of new strategies using available tools, mainly in the areas of genomics and bioinformatics.Citation4 The current review highlights the development of synthetic antigen vaccines, focusing on the usage of bioinformatics tools for the selection and construction of antigens on the different vaccine constructions under development, as well as strategies to optimize vaccines for COVID-19.
Vaccine development landscape in the context of COVID-19
Vaccines are excellent tools in controlling infectious diseases and preventing humanitarian epidemics crisis by inducing the establishment of an immune response capable of quickly controlling and eliminating pathogens. This long-term protection is usually characterized by antibody persistence and cell-mediated immune response.Citation5 As a result, vaccines are the main prophylactic alternative to prevent the spread of COVID-19.Citation6 There are currently 185 candidates being evaluated during the pre-trial vaccine and 102 with eight different technology platforms under clinical evaluationCitation7 (). So far, 17 vaccines have been approved for use in humans in several countries.
Table 1. Main vaccine candidates that are in phases 2/3 of clinical trials or have been approved for emergency use to date
Many laboratories have invested in more modern vaccine strategies besides older vaccine platforms such as the attenuated or inactive virus, especially during the COVID 19 pandemic. A survey carried out in silico by Defrancesco2 showed that several vaccine platforms are being tested, such as protein subunit vaccines, virus-like particle vaccines, DNA- and RNA-based vaccines, viral vector-based vaccines, among other strategies.
Nucleic acid vaccines are new and versatile strategies that use recombinant DNA technology for immunization or immunotherapy. They consist of viral vector-based vaccines, in which a virus unrelated to the pathology, live or inactive, carries the genetic material of the target antigen, along with DNA- and RNA-based vaccine platforms, in which the gene sequence (of one or more genes) encoding the protein of the pathogen of interest will be delivered as a vaccine. Another alternative used in these nucleic acid approaches is the use of epitope coding sequences whose immunogenicity is rigorously selected in silico, in the so-called synthetic antigen vaccine. In this review, we will focus on these synthetic antigen vaccines, which are an interesting strategy since they can combine one or more antigens from the same pathogen or even from different variants in the same vaccine.27,28 In this review, we will focus on DNA and mRNA vaccine platforms, especially multiepitope ones that use synthetic antigens.
Vaccination with non-viral delivered nucleic acid-based approaches has the potential of combining the advantages of live-attenuated vaccine platforms and subunit vaccines, however with no need for cultivation of highly pathogenic organisms on a large scale under biosafety level 3 (BSL3). Furthermore, the inactivation process of viral vaccines can modify the structure of epitopes present in inactivated virus vaccines, which does not occur with nucleic acid approaches. Moreover, because they have no viral particles in their constitution, they do not offer viral reactivation risks, thus providing an excellent option for vulnerable populations, including pregnant women, the elderly, infants, and immunosuppressed people.Citation29 provides a brief description of the different vaccine platforms used against COVID-19 with their advantages and disadvantages.
Another advantage of the next-generation approaches is the much faster and more versatile production of the immunogen. This production makes these platforms ideal for the current chaotic pandemic situation, in which it is necessary to produce billions of doses simultaneously. Another aspect is that, although nucleic acid vaccines have limited coded gene information capacity compared to inactive or attenuated virus vaccines, such synthetic antigens are predicted to be more immunogenic and, because of their reduced size, there is the possibility of combining epitopes from different viral strains in the same vaccine, in addition to working with several vaccine targets simultaneously.
About the flexibility of synthetic antigen vaccines, once the manufacturing process is established, a similar process can be applied to produce a different vaccine by simply replacing the viral antigen coding region with a new insert. Such flexibility makes this vaccine platform ideal for controlling the current pandemic since there is a great possibility of the emergence of new viral variants resistant to the current vaccines in the near future, a situation that requires rapid adaptation of the vaccine. During the construction of synthetic gene, it is possible to evaluate the epitopes conservancy in front of the new coronavirus lineages from the United Kingdom (B.1.1.7),Citation30 South Africa (B. 1.351),Citation30 Brazil (B.1.1.248 – P.1 and P. 2),Citation30,Citation31 India (B.1.617 – B.1.617.1, B.1.617.2 and B.1.617.3),Citation30,Citation32–34 USA (B.1.427 and B.1.429)Citation30,Citation35 and Nigeria (B.1.525),Citation36 as well as its variants. The immunoinformatics tools that work with this analysis will be more detailed in the topic Epitope Conservation analysis.
mRNA vaccines were the first group of platforms approved for emergency use against COVID-19, also representing the platform with the highest levels of effectiveness among all vaccine platforms to date. Although multiepitope vaccines have not registered clinical trials to date, they are still in the immunoinformatics approach phase.Citation37–39
Most candidate vaccines developed to control SARS-CoV-2 infection have the structural antigen S (total length or specific subunits) as their main target. The S glycoproteins are the main responsible for interaction and viral entrance into host cells and based on research on SARS-CoV and MERS-CoV, a strong neutralizing effect was associated to trigger specific cell T responses and neutralizing antibodies, which makes this protein an excellent vaccine target.Citation40,Citation41 Other targets can also be incorporated into multiepitope vaccines, like viral proteins such as E protein, which forms the viral envelope and can be found in higher concentrations during replication of the virus. It can also interact with some cellular proteins and, after the virion construction process, it can break the cell membrane and release the pathogen to the extracellular environmentCitation42,Citation43 which may contribute to the presentation of this antigen to immune system cells. The M protein, in turn, is a membrane protein that is also associated with viral assembly and its specific phosphorylation sites can interact with the host.Citation43,Citation44 While the protein N remains associated with the genetic material of SARS-CoV-2 being related to the viral transcriptional and translational apparatus.Citation43 In addition, Mu et al.Citation45 reported that it can also act in immune system evasion.
Although little explored in studies involving vaccines against COVID-19, accessory proteins can be potential targets for future vaccine constructions based on their importance in the viral construction and how it deals with the immune response from infection. ORF1ab is a polyprotein that is part of the virus replication apparatus. To become functional after entry into the cell, it is cleaved into 11 non-structural proteins that have different functions, being nsp1 known for the possible ability to evade the immune system.Citation43,Citation46 While ORF3a is an ion carrier protein that may be related to the development of the inflammatory process of COVID-19 due to the promotion of cytokine storm besides virulence and viral replication.Citation47–50 ORF6 was considered the protein that showed the highest immunosuppression of primary interferon and its signaling.Citation43 ORF7a, on the other hand, is a protein that acts together with nsp1 and nsp3c in a probable interference in the innate immune response.Citation43,Citation51 Furthermore, mutations in this region should receive greater attention considering that this protein can act as a virulence factor.Citation43 The ORF8, in turn, is a protein that is either related to the pathogenicity or the coronavirus replication apparatus, acting in the interferon pathway of the host. It may also affect the recognition of cytotoxic T lymphocytes by interfering with presentation via MHC and thus evading the immune system,Citation43,Citation52–54 which allows them to explore its use for humoral immune response activation.
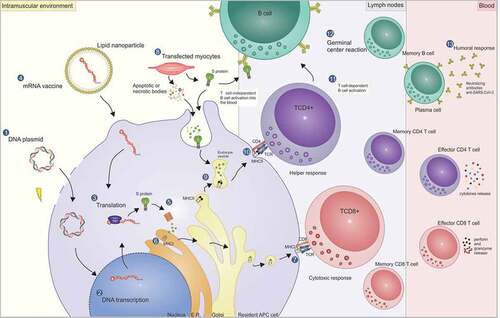
The nucleic acid vaccines can stimulate different arms of the immune response through cross-presentation pathways. The intracellular antigens produced by these vaccines are processed through the endogenous pathway and, therefore, are capable of generating a specific cellular response while still generating antibodies. Besides, synthetic antigen vaccines allow the directioning of immune response by including in the vaccine construct epitopes recognized by B lymphocytes, and MHC-I (cytotoxic response) or MHC-II ligands (helper response). After translation in the cytoplasm, these antigens are generated by proteolysis within the proteasome, followed by their entry into the endoplasmic reticulum via TAP transporter for cell surface presentation. Meanwhile, activation of the helper response occurs via the endocytic pathway, in which somatic cells transfected at the injection site produce the vaccine peptides and these, in turn, can be engulfed by DCs or internalized as apoptotic bodies. Furthermore, such peptides released into the extracellular environment can be directly recognized by B cells or even be presented to these cells via a helper response. More details on all activation pathways generated by nucleic acid approaches, including cellular and humoral responses, can be found in .
Figure 1. Mechanism of action of DNA and mRNA vaccines and the pathways for activating the cellular and humoral response. DNA vaccines are commonly delivered by electroporation through transient pores formed in the membrane (1). Thus, the DNA reaches the cell cytoplasm and then the nucleus, where it will be transcribed (2). Then the mRNA goes to the cytoplasm, where it is translated in the vaccine peptide (3). Another strategy is the direct delivery of the mRNA (mRNA vaccine) encapsulated in lipid nanoparticles in the cell cytoplasm (4). After the endosome escape, the mRNA is translated in the cytoplasm, followed by the vaccine antigen processing in the proteasomes (5), where they are cleaved into smaller peptides. Next, the peptides are transported by the TAP transporter (not shown) into the endoplasmic reticulum, where they are linked to the MHC-I (6) for TCD8 lymphocyte presentation at the cell surface (7), activating the cytotoxic response and generating effective and memory cells. While the cytotoxic response is triggered through the processing of intracellular antigens, the helper response, as a general rule, is triggered through the exogenous pathway, in which transfected somatic cells – such as myocytes at the injection site – produce the vaccine peptide (8). The peptides can be released outside the cell and be directly engulfed by DCs, or they can be internalized by the apoptotic or necrotic bodies, provoked by an inflammatory environment caused by the electroporation. Thus, the fusion of endocytic vesicles – containing the peptides processed by the lysosomal pathway – with vesicles containing MHC-II molecules of DCs (9), allows the presentation of epitopes to the TCD4 lymphocytes at the cell surface (10), with the activation of helper response and generation of memory cells. The TCD4+ lymphocytes, in turn, play a fundamental role in the activation (11) and maturation of B cell affinity inside the germinal centers (12) for the activation of the humoral response (T cell-dependent B cell activation) generating plasmatic cells that can produce high-affinity neutralizing antibodies, as well as memory cells. Another possible activation pathway for humoral response, but with the induction of a weaker immune response, is the direct linkage to the vaccine antigen with B cell receptors (BCRs) (T-cell independent B cell activation).
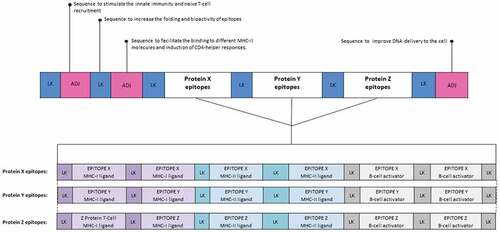
Given the importance of correct processing for the generation and presentation of vaccine epitopes, it is essential to include spacer sequences (also known as linkers) between epitopes in the vaccine construct to provide proteasomal cleavage and TAP binding sites.Citation55 In addition, other linker sequences perform various other functions such as addressing and activating specific routes within cell compartments, more details can be found in . Meanwhile, the schematic representation of a synthetic multi-epitope vaccine construct containing linker sequences can be seen in . Another important in synthetic antigen vaccines is the stability of the antigen after intracellular processing. This analysis is performed using immunoinformatics approaches to each epitope of the vaccine construct. More details of this analysis will be discussed later in the topic of molecular docking analysis and molecular dynamics simulation.
Table 2. Usage of linker sequences in different studies with the aim to ensure the correct processing/directing of peptides in multiepitope vaccines
Figure 2. Structure of a hypothetical synthetic multiepitope vaccine construct containing adjuvant and linkers sequences. In this example, the construct contains sequences that act as adjuvants, which are capable of increasing the immunogenicity of nucleic acid vaccines. Moreover, linker sequences were added between each epitope in order to provide proteasomal and lysosomal processing sites, and TAP transporter binding sites. Concerning the epitopes, in this construction MHC-I, MHC-II ligands, and linear B cell epitopes were added in order to induce both cellular and humoral responses. The epitopes shown in purple are intended for binding to MHC-I molecules and must have between 8 and 11 amino acids. In light blue, the MHC-II ligands are found, these must feature more than 11 amino acids. Meanwhile, the epitopes for B cell activation are shown in gray and contain larger-sized epitopes, up to about 16 aa. LK: Linker, ADJ: Adjuvant.
Development of nucleic acid approaches using immunoinformatics tools
One of the approaches used in the production of genetic vaccines is the usage of Immunoinformatics tools.Citation67 In silico analysis is becoming more important each day, especially because of the pandemic, the lack of financial resources, and the need to construct a vaccine in a short amount of time. Thus, the search for free computational tools became a viable alternative, capable of minimizing the possible limitations that the traditional methods, both in vitro and in vivo, of vaccine construction demand, such as the need for experiments that are not only time-consuming but need a good laboratory infrastructure, which is very expensive.
Advancements in bioinformatics contributed to the development of new tools for the analysis of protein compounds with drug potential and the assistance in vaccine construction.Citation67 It was noted that after the first SARS-CoV-2 genetic sequence was deposited in GenBank,Citation68 many studies were able to use these computational tools during the pandemics.Citation4,Citation69–75 Therefore, it is possible to believe that immunologic bioinformatics tools, also named immunoinformatics approaches, tend to grow even more after the pandemic.
In silico analysis encompasses a wide range of production steps for a gene vaccine against COVID-19, such as the prediction of epitopes, immunogenicity and conservation analysis, populational coverage evaluation, molecular docking, and molecular dynamics simulation of the epitope-MHC complex.Citation76 These analyses allow the selection of epitopes that potentially are more effective,Citation77 which is less time-consuming when compared to in vitro screening.
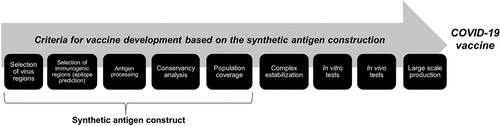
It is possible to build a synthetic multiepitope gene that will be further validated in vitro and in vivo in order to be used in the vaccine trials (). This synthetic gene, when transcribed and translated by cells, will act as a synthetic antigen, which will hopefully be recognized by the immune system, activate the T and B lymphocytes, and produce antibodies.Citation78 Following, there is a list of steps and tools used in the construction of a synthetic antigen.
Figure 3. Summary showing, step by step, the criteria for the development of a COVID-19 vaccine through the construction of synthetic antigens.
Epitope prediction
The GenBank (https://www.ncbi.nlm.nih.gov/genbank/), at the National Center for Biotechnology Information (NCBI), is a database of genetic sequences known worldwide, where nucleotide sequences for a wide range of organisms can be found. In addition, NCBI has a database for amino acid sequences, the Protein Database (https://www.ncbi.nlm.nih.gov/protein). The availability of amino acid sequences for each protein of the new coronavirus enables the prediction of epitopes. This step is fundamental to the construction of a synthetic antigen that can be used in nucleic acid approaches against COVID-19 because it corresponds to the selection of peptides from virus proteins that could bind to MHC (major histocompatibility complex) molecules capable of inducing T (CD8+ and CD4+) and B cells activation.
The predictions can be carried out through different computational methods, such as Artificial Neural Networks, NetMHCpan, Stabilized Matrix Method (SMM), Matrix Vector Support (SVM), NetCL/NetCLpan/NetCHOP, Consensus,Citation79–85 among others (). Those methods are used in different databases and online servers, such as the Immune Epitope Database and Analysis Resource (IEDB) (https://www.iedb.org/), Virus Pathogen Resource (ViPR) (https://www.viprbrc.org/), NetCTLpan – 1.1,Citation90 NetMHCpan – 4.0,Citation80 NetMHCstab – 1.0,Citation92 NetMHCstabpan – 1.0,Citation93 NetCTL 1.2,Citation94 ProPred-I and ProPred,Citation95,Citation96 RANKPEP,Citation97 among others.
Table 3. In silico methods to predict T cells epitopes
Some of these tools and servers are already used in COVID-19 research, such as Abdelmageed et al.,Citation71 Rahman et al.,Citation75 and Dong et al.,Citation59 who have used the IEDB tools to select T cell epitopes, Ahmed et al.Citation70 used ViPR to predict T and B cell epitopes, Bhattacharya et al.Citation73 used the ProPred-I and ProPred to predict MHC-I and MHC-II linker epitopes, Grifoni et al.Citation98 did the epitope prediction of MHC-II using the NetMHCpan EL – 4.0 server, and Enayatkhani et al.Citation74 predicted MHC-I and MHC-II epitopes using the RANKPEP server in order to design a multiepitope vaccine against COVID-19.
These immunoinformatics tools available in the databases and servers demand that the type of human MHCs (HLAs) of interest is informed, so it can provide the epitopes for T CD8+ and CD4 + . For vaccines against COVID-19,Citation99 a list of HLAs with high affinity to SARS-CoV-2 peptides was made available, displaying the worldwide amplitude that can be used in prediction tools. Some of the alleles that present a strong binding with these peptides were HLA-A*02:11, HLA-A*02:22, HLA-A*02:02, HLA-A*02:03, HLA-A*02:06, HLA-B*15:03, HLA-B*15:17, HLA-B*35:10, HLA-B*15:25, HLA-B*15:39, HLA-C*03:02, HLA-DRB1*01:01, HLA-DRB1*10:01, HLA-DRB1*01:04, HLA-DRB1*11:02, HLA-DRB1*13:01. All these alleles were capable of binding to more than 100 peptides. Besides these, other HLAs ligands to SARS-CoV-2 can be found in the consortium formed during the pandemics, named COVID-19 HLA & Immunogenetics (http://www.hlacovid19.org/), which has a specific database for those who work with COVID-19. Another database containing HLAs of different populations worldwide is the Allele Frequency Net Database,Citation100 which was used by Moura et al.Citation76 to identify epitopes in the S protein of SARS-CoV-2.
According to the processing of peptides by the cell proteasome, the efficiency of its displacement by the TAP channel, and the binding capacity to HLAs molecules, it is possible to detect potential epitopes.Citation82 The NetChop-3.1 serverCitation89 detects the peptide from the proteasomal cleavage sites, while the MHC I processing tool (Proteasome, TAP)Citation84 was used in the in silico design for the COVID-19 vaccine from S, M, and E proteins done by Rahman et al.,Citation75 which generates a ranking based on the potential of each T cell epitope.
The peptides that have a higher potential to be considered a T cell epitope must go through an immunogenicity analysis since not all peptides are immunogenic.Citation101 This analysis generally consists of an evaluation of the peptide capacity of inducing lymphocyte activation. It can be done using a tool available in the IEDB named Class I Immunogenicity,Citation102 as suggested by Kardani et al.,Citation103 or the C-ImmSim server,Citation104 as used by Dong et al.Citation59 for the construction of in silico multiepitope vaccine against COVID-19. It can also be done through the NETMHCpan – 4.0 server,Citation80 which was used by Moura et al.Citation76
The general method for the prediction of B cells is based on the residual value and the informed quantity of amino acids around the residue. The amino acid amplitude capable of defining a peptide that has the antigenic potential varies between 5 and 7 amino acids. Rahman et al.Citation75 performed this analysis in their coronavirus studies using the ABCPred serversCitation105 and BepiPred-2.0.Citation106 The same methods are also available in the IEDB database, the Antibody Epitope Prediction (http://tools.iedb.org/bcell/), which was used by Bhattacharya et al.Citation73 and Grifoni et al.Citation98 The prediction tool available in the Virus Pathogen Resource (ViPR) (https://www.viprbrc.org/) was used in the SARS-CoV-2 study done by Ahmed et al.Citation70
From the predicted epitopes it is possible to identify their antigenic potential. In studies related to COVID-19, such as the ones done by Baruah and Bose,Citation72 Bhattacharya et al.,Citation73 Dong et al.,Citation59 Enayatkhani et al.Citation74 and Rahman et al.,Citation75 the antigenicity analysis was done through the VaxiJen server.Citation107
Epitope clusters
It is possible to have sequence similarities among the predicted epitopes, thus allowing for clusters to be created. Clusters are groups that unite the epitopes that were predicted over the same regions. This step avoids information redundancy regarding the same epitope. The Epitope Cluster AnalysisCitation108 can be used in the design for the vaccine against COVID-19, focusing on cluster identification, which is available at the IEDB. This tool gathers epitopes that have over 80% similarity and defines the epitopes represented in each cluster. EpiMatrix and ClustiMer are also servers capable of identifying epitope clusters that can be used in vaccine constructions, as observed in the study of Scholzen et al.Citation109
Epitope conservation analysis
Among the virus protein variants, the predicted epitopes can be conserved or not. Thus, in order to have a vaccine that prolonged immunity even when faced with different variants, it is important to verify the level of conservation of these epitopes and select those that have higher conservation levels.Citation103 The Epitope Conservancy Analysis tool,Citation110 available at the IEDB, can be used to identify the more conserved epitopes of T and B cells to be added in the multiepitope construction against SARS-CoV-2. This tool calculates a value referring to the level of conservation from a certain level of identity (obtained by the analysis of epitope clusters) and defines a ranking from the generated values.
Populational coverage analysis
Considering the importance of a vaccine capable of covering most of the population for containing the pandemic, it is vital to perform an analysis of the populational coverage. This analysis will verify the populations around the world and check for common alleles capable of interacting with the epitopes. Kardani et al.Citation103 mentioned different tools for in silico vaccine design against different pathogen microorganisms, amongst which is SARS-CoV-2, reporting the use of Population Coverage tool,Citation111 available at the IEDB. Abdelmageed et al.Citation71 and Rahman et al.Citation75 also used this tool to analyze the population coverage of predicted epitopes. This tool calculates the coverage fractions of HLAs for the populations.
The best results found in this phase can define whether more than one vaccine will need to be designed. Kibria et al.Citation112 demonstrated the importance of this analysis when they realized that it would be needed to design two vaccines at the end of the study because one of the epitopes predicted presented low coverage for the South African population (3.15%) when compared to another predicted epitope (40.9%). Therefore, it was necessary to design a vaccine exclusive for the South African population and another for the rest of the populations worldwide.
Molecular docking analysis and molecular dynamics simulation
The epitopes that presented higher populational coverage values can be used in a molecular docking analysis. The docking is performed to calculate the best pose and the binding energy between the predicted epitopes and MHC molecules. ClusPro,Citation113 PatchDock,Citation114 HADDOCK 2.4,Citation115 AutoDock 4.0 (http://autodock.scripps.edu/), CABS-dockCitation116 and ZDOCK 3.0.2Citation117 are some of the online servers used in many studies about COVID-19.Citation59,Citation71–76
For this kind of molecular docking, it is necessary to use 3D structures of the HLAs available at the Protein Data Bank (PDB) (https://www.rcsb.org/). Intending to aid COVID-19 studies, the PDB has a section exclusively for SARS-CoV-2 structures. Bhattacharya et al.Citation73 used in their study for the design of a vaccine against the new coronavirus, the file with the docked complex so it can be visualized in PyMOL software (https://pymol.org/2/).
With the complexes formed with the peptides bound to HLA molecules, it is possible to perform a molecular dynamics (MD) simulation. This analysis assesses the stability of the peptide-HLA complex through a certain amount of time under specific temperature, pressure, ion presence, and water molecule conditions, simulating the conditions of the biological process related to the peptide-HLA binding complex. For that, the complex needs to remain stable during enough time for lymphocyte activation.Citation118
NAMD (https://www.ks.uiuc.edu/Research/namd/) is one of the programs that performs molecular dynamics simulation, and the Visual Molecular Dynamics (VMD) program can be used to visualize its results (https://www.ks.uiuc.edu/Research/vmd/). Baruah and BoseCitation72 used these programs to perform an MD simulation to assess the stability of the complex peptide-MHC of T and B cells of glycoproteins on the surface of the new coronavirus. Dong et al.Citation59 used the server GROMACS (http://www.gromacs.org/) for MD simulations in their multiepitope vaccine constructions against COVID-19.
Reverse translation and synthetic antigen production
After filtering the epitopes that present higher stability in MD simulations, the amino acid sequences can be back translated into nucleotides, so a synthetic gene can be constructed. The Reverse Translate programCitation119 allows the back translation of amino acid sequences into nucleotides. These sequences, when put together, form a bigger sequence composed of nucleotides capable of synthesizing all selected epitopes. Therefore, it is possible to insert it into a plasmid vector, for example, configuring a gene vaccine. When it enters the organism, the body recognizes it as a synthetic antigen and activates the immune system, providing the necessary response to protect the person who was vaccinated.Citation78
This construction step of the candidate vaccine structure against SARS-CoV-2 was possible to be observed in the study of Enayatkhani et al.Citation74 who constructed the secondary structure of the vaccine using the server PSIPRED (http://bioinf.cs.ucl.ac.uk/web_servers/psipred_server/psipred_overview/) and in silico cloned it using the SnapGene software (https://www.snapgene.com/). Dong et al.Citation59 opted to use the JCat toolCitation120 to design their multiepitope vaccine against COVID-19.
The use of different computational tools for the prediction and analysis of epitopes allows that only virtually the best epitopes are selected, with the best results of immunogenicity, conservation, populational coverage, binding energy, and stability. Therefore, these filters can make vector-based approaches faster and more efficient.Citation121
DNA and RNA based vaccines are essentially poorly immunogenic,Citation122 thus, the administration of adjuvants is essential to overcome this limitation.Citation123 An important class of adjuvants are Toll-like receptors (TLR) ligands. When stimulated, the TLR rapidly identify these molecules as “dangerous” and trigger the production of pro-inflammatory cytokines, as well as the activation of innate immune response, and the increase of antigen presentation to lymphocytes by dendritic cells (DCs). Examples of TLRs agonist are the TLR-9 agonist composed of CpG motifs, which are capable of inducing a strong cytotoxic responseCitation124 and the TLR-3 agonist molecule polyriboinosinic polyribocytidylic acid [Poly(I:C)], which is a double-stranded RNA analogue capable of inducing cell signaling through multiple inflammatory pathways.Citation125,Citation126 Another promising class of immunomodulator are cytokines, since these proteins play a critical role in immune cell signaling. Several studies have included plasmids encoding cytokines in their assays,Citation127 such as the use of IL-2 and IL-12 in vaccines for influenza,Citation128 SARS-CoV,Citation129 and HIVCitation130–132 which demonstrated the significant increase of immunogenicity. Finally, it is essential to ensure the efficiency of the vaccine transfection, so the most promising delivery systems for nucleic acid approaches include electroporation (EP) for DNA-based vaccines and lipid nanoparticles (LNPs) for mRNA vaccines, resulting in increased uptake of the vaccine plasmid and consequently increasing its efficiency.Citation133–135
Conclusion
The COVID-19 pandemic brought to light that viral diseases have the potential of decimating millions of people in a short amount of time, something that happened before until efficient vaccines were developed that allowed the control of these diseases. Such vaccines were developed by classic platforms that contributed to major advances in public health, such as the eradication of smallpox. However, certain limitations are associated with these platforms, which make them less susceptible to the rapid response that a pandemic requires. We are currently facing an unprecedented effort at accelerated speed during vaccine development, in which numerous research groups worldwide have been working simultaneously, along with governmental and private efforts to try to curb the infection.
The enormous advances in molecular engineering and biotechnology in recent decades have enabled the development of increasingly efficient bioactive molecules, such as the latest generation vaccines. Such vaccine platforms have numerous advantages, such as greater safety; better immune response directioning; the possibility of coverage against multiple viral subtypes; the fast development, production, and ease of storage, which justifies the growing effort to establish these vaccine strategies. Additionally, the databases and the bioinformatics tools currently available allow the prediction of the most promising epitopes to use in essays in vivo, also allowing rapid replacement of these epitopes in other vaccine constructs in response to pathogen mutations, thus preventing epidemics with emerging viral subtypes.
The current pandemic context is surrounded by challenges. One of them is the development in record time of a vaccine for a new virus in which it is still spreading at alarming rates and constantly mutating, in which there is a need for the production and distribution of billions of doses. In addition, the immunopathogenesis of COVID-19 is not fully understood, and previous studies from vaccines against the following viruses (SARS-CoV and MERS-CoV) in some animal models raised safety concerns regarding Th2 mediated immunopathology.Citation136
Another challenge is the reconsideration of current approaches to regulatory assessment and the licensing process of new vaccine platforms by government agencies in order to ensure the safety and efficacy of these new vaccines, which is a time-consuming factor. However, time is a crucial element in the current context, since the SARS-CoV-2 virus reached an average worldwide infection rate of 828 thousand people a day and 14,7 thousand deaths during the peak of the pandemic (to date). History showed us that these crises also generate unique opportunities for the development of new technologies, and it is possible that the learning generated with SARS-CoV-2 will revolutionize vaccine development technology for human usage, which is already proving to be highly effective and safe, and therefore, this can open the field to many possibilities that are not restricted to prophylactic but also therapeutic purposes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- World Health Organization (WHO). Coronavirus disease (COVID-19) dashboard. Geneva, Switzerland: World Health Organization; acessed 2021 Jul 31. https://covid19.who.int/.
- DeFrancesco L. Whither COVID-19 vaccines? Nat Biotechnol. 2020. [acessed 2021 Mar 2]. https://www.nature.com/articles/s41587-020-0697-7#citeas.
- World Health Organization (WHO). Global research on coronavírus disease (COVID-19). Geneva, Switzerland: World Health Organization [acessed 2021 Feb 3]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov.
- Florindo HF, Kleiner R, Vaskovich-Koubi D, Acúrcio RC, Carreira B, Yeini E, Tiram G, Liubomirski Y, Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat Nanotechnol. 2020;15(8):630–45. doi:10.1038/s41565-020-0732-3.
- Bernasconi V, Kristiansen PA, Whelan M, Román RG, Bettis A, Yimer SA, Gurry C, Andersen SR, Yeskey D, Mandi H, et al. Entwicklung von Impfstoffen gegen neu auftretende Infektionskrankheiten mit epidemischem Potenzial. Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz. 2020;63(1):65–73. doi:10.1002/prot.21078.
- Chen W-H, Strych U, Hotez PJ, Bottazzi ME. The SARS-CoV-2 vaccine pipeline: an overview. Current Tropical Medicine Reports. 2020;7(2):61–64. doi:10.1007/s40475-020-00201-6.
- World Health Organization (WHO). Draft landscape and tracker of COVID-19 candidate vaccines. Geneva, Switzerland: World Health Organization [acessed 2021 Mar 21]. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- Palacios R, Patiño EG, de Oliveira Piorelli R, Conde MTRP, Batista AP, Zeng G, Xin Q, Kallas EG, Flores J, Ockenhouse CF, et al. Double-blind, randomized, placebo-controlled Phase III clinical trial to evaluate the efficacy and safety of treating healthcare professionals with the adsorbed COVID-19 (Inactivated) vaccine manufactured by Sinovac – PROFISCOV: a structured summary of a. Trials. 2020;21:1–3. doi:10.1186/s13063-020-04775-4.
- Xia S, Duan K, Zhang Y, Zhao D, Zhang H, Xie Z, Li X, Peng C, Zhang Y, Zhang W, et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes. JAMA. 2020;324:951–60. doi:10.1001/jama.2020.15543.
- Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, Xu W, Zhao Y, Li N, Zhang J, et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–21. doi:10.1016/j.cell.2020.06.008.
- Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, Bellamy D, Bibi S, Bittaye M, Clutterbuck EA, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–78. doi:10.1016/S0140-6736(20)31604-4.
- Barrett JR, Belij-Rammerstorfer S, Dold C, Ewer KJ, Folegatti PM, Gilbride C, Halkerston R, Hill J, Jenkin D, Stockdale L, et al. Phase 1/2 trial of SARS-CoV-2 vaccine ChAdOx1 nCoV-19 with a booster dose induces multifunctional antibody responses. Nat Med. 2021;27:279–88. doi:10.1038/s41591-020-01179-4.
- Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet. 2021;397:99–111. doi:10.1016/S0140-6736(20)32661-1.
- Zhu F-C, Li Y-H, Guan X-H, Hou L-H, Wang W-J, Li J-X, Wu S-P, Wang B-S, Wang Z, Wang L, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–54. doi:10.1016/S0140-6736(20)31208-3.
- Zhu F-C, Guan X-H, Li Y-H, Huang J-Y, Jiang T, Hou L-H, Li J-X, Yang B-F, Wang L, Wang W-J, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–88. doi:10.1016/S0140-6736(20)31605-6.
- Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatulin AI, Shcheblyakov DV, Dzharullaeva AS, Grousova DM, Erokhova AS, Kovyrshina AV, Botikov AG, et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–97. doi:10.1016/S0140-6736(20)31866-3.
- Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, Kovyrshina AV, Lubenets NL, Grousova DM, Erokhova AS, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–81. doi:10.1016/S0140-6736(21)00234-8.
- Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, Groot AM, Stoop J, Tete S, Damme WV, Leroux-Roels I, et al. Safety and immunogenicity of the Ad26.COV2.S COVID-19 vaccine candidate: interim results of a phase 1/2a, double-blind, randomized, placebo-controlled trial. MedRxiv. 2020;1–28. doi:10.1101/2020.09.23.20199604.
- Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, Plested JS, Zhu M, Cloney-Clarck S, Zhou H, et al. Phase 1–2 Trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med. 2020;383:2320–32. doi:10.1056/NEJMoa2026920.
- Tebas P, Yang S, Boyer JD, Reuschel EL, Patel A, Christensen-Quick A, Andrade VM, Morrow MP, Kraynyak K, Agnes J, et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: a preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine. 2021;31:1–9. doi:10.1016/j.eclinm.2020.100689.
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chapell JD, Denison MR, Stevens LJ, et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N Engl J Med. 2020;383:1920–31. doi:10.1056/NEJMoa2022483.
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–16. doi:10.1056/NEJMoa2035389.
- Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–93. doi:10.1038/s41586-020-2639-4.
- Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, Baum A, Pascal K, Quandt J, Maurus D. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–99. doi:10.1038/s41586-020-2814-7.
- Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, et al. Safety and Immunogenicity of Two RNA-based COVID-19 vaccine candidates. N Engl J Med. 2020;383(25):2439–50. doi:10.1056/NEJMoa2027906.
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Marc GP, Moreira ED, Zerbini C, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–15. doi:10.1056/NEJMoa2034577.
- Maruggi G, Zhang C, Li J, Ulmer JB, Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol Ther. 2019;27(4):757–72. doi:10.1016/j.ymthe.2019.01.020.
- Chahal JS, Khan OF, Cooper CL, McPartlan JS, Tsosie JK, Tilley LD, Sidik SM, Lourido S, Langer R, Bavari S, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci USA. 2016;113(29):E4133–4142. doi:10.1073/pnas.1600299113.
- Carter C, Houser KV, Yamshchikov GV, Bellamy AR, May J, Enama ME, Sarwar U, Larkin B, Bailer RT, Koup R, et al. Safety and immunogenicity of investigational seasonal influenza hemagglutinin DNA vaccine followed by trivalent inactivated vaccine administered intradermally or intramuscularly in healthy adults: an open-label randomized phase 1 clinical trial. PLoS One. 2019;14(9):1–18. doi:10.1371/journal.pone.0222178.
- Gupta RKW. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nature Rev Immunol. 2021;21(6):340–41. doi:10.1038/s41577-021-00556-5.
- Toovey O, Harvey KN, Bird PW, Tang J. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J Infect. 2021;82(5):e23–e24. doi:10.1016/j.jinf.2021.01.025.
- Garcia V, Vig V, Peillard L, Ramdani A, Mohamed S, Halfon P. First description of two immune escape indian B. 1.1. 420 and B. 1.617. 1 SARS-CoV2 variants in France. bioRxiv. 2021:1–9. doi:10.1101/2021.05.12.443357.
- Eales O, Page A, Tang SN, Walters CE, Wang H, Haw D, Trotter AJ, Viet TL, Foster-Nyarko E, Prosolek S, et al. SARS-CoV-2 lineage dynamics in England from January to March 2021 inferred from representative community samples. medRxiv. 2021;1–33. doi:10.1101/2021.05.08.21256867.
- Hutchinson D, Williams H, Stone H. COVID-19 variants of concern in Australia, September 2020-April 2021. Global Biosecurity. 2021;3:1–22. doi:10.31646/gbio.111.
- Webb LM, Matzinger S, Grano C, Kawasaki B, Stringer G, Bankers L, Herlihy R. Identification of and Surveillance for the SARS-CoV-2 Variants B.1.427 and B.1.429 — Colorado, January–March 2021. MMWR Morb Mortal Wkly Rep. 2021;70(19):717–18. doi:10.15585/mmwr.mm7019e2.
- Ozer EA, Simons LM, Adewumi OM, Fowotade AA, Omoruyi EC, Adeniji JA, Dean TJ, Taiwo BO, Hultquist JF, Lorenzo-Redondo R. High prevalence of SARS-CoV-2 B. 1.1. 7 (UK variant) and the novel B. 1.5. 2.5 lineage in Oyo State, Nigeria. medRxiv. 2021:1–22. doi:10.1101/2021.04.09.21255206.
- Khan A, Khan S, Saleem S, Nizam-Uddin N, Mohammad A, Khan T, Ahmad S, Arshad M, Ali SS, Suleman M, et al. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput Biol Med. 2021;133:1–11. doi:10.1016/j.compbiomed.2021.104420.
- Ita K. Coronavirus disease (COVID-19): current status and prospects for drug and vaccine development. Arch Med Res. 2021;52(1):15–24. doi:10.1016/j.arcmed.2020.09.010.
- Rajput VS, Sharma R, Kumari A, Vyas N, Prajapati V, Grover A. Engineering a multi epitope vaccine against SARS-CoV-2 by exploiting its non structural and structural proteins. J Biomol Struct Dyn. 2021:1–18. doi:10.1080/07391102.2021.1924265.
- Dagotto G, Yu J, Barouch DH. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe. 2020;28(3):364–70. doi:10.1016/j.chom.2020.08.002.
- Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):1–28. doi:10.3390/v11010059.
- Kuo L, Hurst KR, Masters PS. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. Journal of Virology. 2007;81(5):2249–62. doi:10.1128/JVI.01577-06.
- Timmers L, Peixoto J, Ducati R, Bachega JF, Pereira LM, Caceres RA, Majolo F, Da Silva GL, Anton DB, Goettert MI, et al. SARS-CoV-2 mutations in Brazil: from genomics to clinical conditions. Chemrxiv. 2021;1–32. doi:10.26434/chemrxiv.14045783.v1.
- Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Marrero MC, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM, et al. The global phosphorylation landscape of SARS-CoV-2. Infection Cell. 2020;8674:30811–14. doi:10.1016/j.cell.2020.06.034.
- Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M, Ren Y, Li X, Geng Q, Xu Y, et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci. 2020;63(9):1413–16. doi:10.1007/s11427-020-1692-1.
- Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–55. doi:10.1126/science.abc8665.
- Tan Y-J, Tham P-Y, Chan DZL, Chou C-F, Shen S, Fielding BC, Tan THP, Lim SG, Hong W. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J Virol. 2005;79:10083–87. doi:10.1128/JVI.79.15.10083-10087.2005.
- Issa E, Merhi G, Panossian B, Salloum T, Tokajiana S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems. 2020;5(3):e00266–20. doi:10.1371/journal.pcbi.1003266.
- Ren Y, Shu T, Wu D, Mu J, Wang C, Huang M, Han Y, Zhang X-Y, Zhou W, Qiu Y, et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol. 2020;17(8):881–83. doi:10.1038/s41423-020-0485-9.
- Siu K-L, Yuen K-S, Castano‐Rodriguez C, Ye Z-W, Yeung M-L, Fung S-Y, Yuan S, Chan C-P, Yuen K-Y, Enjuanes L, et al. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33(8):8865–77. doi:10.1096/fj.201802418R.
- Taylor JK, Coleman CM, Postel S, Sisk JM, Bernbaum JG, Venkataraman T, Sundberg EJ, Frieman MB. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. Journal of Virology. 2015;89(23):467–78. doi:10.1016/S0140-6736(20)31604-4.
- Khailanya RA, Safdarb M, Ozaslanc M. Genomic characterization of a novel SARS-CoV- 2. Gene Rep. 2020;19:1–6. doi:10.1016/j.genrep.2020.100682.
- Jin-Yan L, Ce-Heng L, Qiong W, Yong-Jun T, Rui L, Ye Q, Xing-Yi G. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:1–11. doi:10.1016/j.virusres.2020.198074.
- Zhang Y, Zhang J, Chen Y, Luo B, Yuan Y, Huang F, Yang T, Yu F, Liu J, Liu B, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. Nat Acad Sci. 2021;118:1–12. doi:10.1073/pnas.2024202118.
- Livingston B, Crimi C, Newman M, Higashimoto Y, Appella E, Sidney J, Sette A. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J Immunol. 2002;168(11):5499–506. doi:10.4049/jimmunol.168.11.5499.
- Reguzova A, Antonets D, Karpenko L, Ilyichev A, Maksyutov R, Bazhan S. Design and evaluation of optimized artificial HIV-1 poly-T cell-epitope immunogens. PLoS One. 2015;10(3):1–18. doi:10.2144/00286ir01.
- Bazhan SI, Karpenko LI, Ilyicheva TN, Belavin PA, Seregin SV, Danilyuk NK, Antonets DV, Ilyichev AA. Rational design based synthetic polyepitope DNA vaccine for eliciting HIV-specific CD8+ T cell responses. Mol Immunol. 2010;47(7–8):279–88. doi:10.1038/s41591-020-01179-4.
- Wang Q-M, Sun S-H, Hu Z-L, Zhou F-J, Yin M, Xiao C-J, Zhang J-C. Epitope DNA vaccines against tuberculosis: spacers and ubiquitin modulates cellular immune responses elicited by epitope DNA vaccine. Scand J Immunol. 2004;60(3):W526–W531. doi:10.1093/nar/gki376.
- Dong R, Chu Z, Yu F, Zha Y. Contriving multi-epitope subunit of vaccine for COVID-19: immunoinformatics approaches. Front Immunol. 2020;11:1–18. doi:10.3389/fimmu.2020.01784.
- Onile OS, Ojo GJ, Oyeyemi BF, Agbowuro GO, Fadahunsi AI. Development of multiepitope subunit protein vaccines against Toxoplasma gondii using an immunoinformatics approach. NAR Genomics Bioinforma. 2020;2:48. doi:10.1093/nargab/lqaa048.
- Liu H, Shen W, Shu J, Kou Z, Jin X. A novel polyepitope vaccine elicited HIV peptide specific CD4+ T cell responses in HLA-A2/DRB1 transgenic mice. PLoS One. 2017;12:1–11. doi:10.1371/journal.pone.0184207.
- Nezafat N, Ghasemi Y, Javadi G, Khoshnoud MJ, Omidinia E. A novel multi-epitope peptide vaccine against cancer: an in silico approach. J Theor Biol. 2014;349:121–34. doi:10.1016/j.jtbi.2014.01.018.
- Levy A, Pitcovski J, Frankenburg S, Elias O, Altuvia Y, Margalit H, Peretz T, Golenser J, Lotem M, Zhu F-C, et al. A melanoma multiepitope polypeptide induces specific CD8+ T-cell response. Cell Immunol. 2007;250(1–2):24–30. doi:10.1016/S0140-6736(20)31605-6.
- Cardinaud S, Bouziat R, Rohrlich P-S, Tourdot S, Weiss L, Langlade-Demoyen P, Burgevin A, Fiorentino S, Endert PV, Lemonnier FA. Design of a HIV-1-derived HLA-B07.02-restricted polyepitope construct. AIDS. 2009;23(15):1945–54. doi:10.1097/QAD.0b013e32832fae88.
- Schneider SC, Ohmen J, Fosdick L, Gladstone B, Guo J, Ametani A, Sercarz EE, Deng H. Cutting edge: introduction of an endopeptidase cleavage motif into a determinant flanking region of hen egg lysozyme results in enhanced T cell determinant display. J Immunol. 2000;165(1):20–23. doi:10.1056/NEJMoa2026920.
- Ji H, Wang T-L, Chen C-H, Pai SI, Hung C-F, Lin K-Y, Kurman RJ, Pardoll DM, Wu TC. Targeting human papillomavirus Type 16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine human papillomavirus Type 16 E7-expressing tumors. Hum Gene Ther. 1999;10(17):2727–40. doi:10.1089/10430349950016474.
- Verma S, Sajid A, Singh Y, Shukla P. Computational tools for modern vaccine development. Hum Vaccines Immunother. 2020;16(3):723–35. doi:10.1080/21645515.2019.1670035.
- Wu F, Wang A, Liu M. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020;00:1–24. doi:10.2139/ssrn.3566211.
- Qamar MTU, Rehman A, Tusleem K, Ashfaq UA, Qasim M, Zhu X, Fatima I, Shahid F, Chen -L-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: immunoinformatics and in silico approaches. PLoS One. 2020;15:1–25. doi:10.1371/journal.pone.024417.
- Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV immunological studies. Viruses. 2020;12:1–15. doi:10.3390/v12030254.
- Abdelmageed MI, Abdelmoneim AH, Mustafa MI, Elfadol NM, Murshed NS, Shantier SW, Makhawi AM. Design of a multiepitope-based peptide vaccine against the e protein of human COVID-19: an immunoinformatics approach. Biomed Res Int. 2020;00:1–12. doi:10.1155/2020/2683286.
- Baruah V, Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol. 2020;92:495–500. doi:10.1002/jmv.25698.
- Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee S-S CC. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol. 2020;92:618–31. doi:10.1002/jmv.25736.
- Enayatkhani M, Hasaniazad M, Faezi S, Gouklani H, Davoodian P, Ahmadi N, Einakian MA, Karmostaji A, Ahmadi K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J Biomol Struct Dyn. 2020;2:1–16. doi:10.1080/07391102.2020.1756411.
- Rahman MS, Hoque MN, Islam MR, Akter S, Alam ASMRU, Siddique MA, Saha O, Rahaman MM, Sultana M, Crandall KA, et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. PeerJ. 2020;8:1–30. doi:10.7717/peerj.9572.
- Moura RR, Agrelli A, Santos-Silva CA, Silva N, Assunção BR, Brandão L, Benko-Iseppon AM, Crovella S. Immunoinformatic approach to assess SARS-CoV-2 protein S epitopes recognised by the most frequent MHC-I alleles in the Brazilian population. J Clin Pathol. 2020;00:1–5. doi:10.1136/jclinpath-2020-206946.
- Khuroo MS, Khuroo M, Khuroo MS, Sofi AA, Khuroo NS. COVID-19 vaccines: a race against time in the middle of death and devastation! J Clin Exp Hepatol. 2020;10:610–21. doi:10.1016/j.jceh.2020.06.003.
- Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines. 2012;11:189–209. doi:10.1586/erv.11.188.
- Desai DV, Kulkarni-Kale U. T-cell epitope prediction methods: an overview. In: De R, Tomar N, editors. Immunoinformatics. methods in molecular biology (Methods and Protocols). New York (NY): Humana Press; 2014. p. 333–64.
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved peptide–MHC Class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199:3360–68. doi:10.4049/jimmunol.1700893.
- Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform. 2005;6:1–9. doi:10.1186/1471-2105-6-132.
- Peters B, Bulik S, Tampe R, Van Endert PM, Holzhutter HG. Identifying MHC Class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–49. doi:10.4049/jimmunol.171.4.1741.
- Peters B, Tong W, Sidney J, Sette A, Weng Z. Examining the independent binding assumption for binding of peptide epitopes to MHC-I molecules. Bioinformatics. 2003;19:1765–72. doi:10.1093/bioinformatics/btg247.
- Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel PM, Rammensee HG, Schild H, Holzhutter HG. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci. 2005;62:1025–37. doi:10.1007/s00018-005-4528-2.
- Wang S-C. Artificial neural network (ANNs). In: interdisciplinary computing in Java programming. Springer,Boston. 2003;743:81–100. doi:10.1007/978-1-4615-0377-4_5.
- Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform. 2005;6:1–9. doi:10.1186/1471-2105-6-132.
- Keşmir C, Nussbaum AK, Schild H, Detours V, Brunak S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 2002;15:287–96. doi:10.1093/protein/15.4.287.
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, Lund O, Nielsen M. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35:2295–303. doi:10.1002/eji.200425811.
- Nielsen M, Lundegaard C, Lund O, Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi:10.1007/s00251-005-0781-7.
- Stranzl T, Larsen MV, Lundegaard C, Nielsen M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics. 2010;62:357–68. doi:10.1007/s00251-010-0441-4.
- Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:1–10. doi:10.1371/journal.pcbi.1000048.
- Jørgensen KW, Rasmussen M, Buus S, Nielsen M. NetMHCstab - Predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery. Immunology. 2014;141:18–26. doi:10.1111/imm.12160.
- Rasmussen M, Fenoy E, Harndahl M, Kristensen AB, Nielsen IK, Nielsen M, Buss S. Pan-specific prediction of peptide–MHC Class I complex stability, a correlate of T cell immunogenicity. J Immunol. 2016;197:1517–24. doi:10.4049/jimmunol.1600582.
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8:1–12. doi:10.1186/1471-2105-8-424.
- Singh H, Raghava GPS. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–37. doi:10.1093/bioinformatics/17.12.1236.
- Singh H, Raghava GPS. ProPred1: prediction of promiscuous MHC class-I binding sites. Bioinformatics. 2003;19:1009–14. doi:10.1093/bioinformatics/btg108.
- Reche PA, Reinherz EL. Prediction of peptide-MHC binding using profiles. Methods Mol Biol. 2007;409:185–200. doi:10.1007/978-1-60327-118-9_13.
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, et al. Targets of T cell responses to SARS-CoV-2 Coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–501. doi:10.1016/j.cell.2020.05.015.
- Barquera R, Collen E, Di D, Buhler S, Teixeira J, Llamas B, Nunes JM, Sanchez‐Mazas A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020;96:277–98. doi:10.1111/tan.13956.
- Gonzalez-Galarza FF, McCabe A, Santos EJM, Jones J, Takeshita L, Ortega-Rivera ND, Cid-Pavon GMD, Ramsbottom K, Ghattaoraya G, Alfirevic A, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48:783–88. doi:10.1093/nar/gkz1029.
- Feltkamp MCW, Vierboom MPM, Kast WM, Melief CJM. Efficient MHC class I-peptide binding is required but does not ensure MHC class I-restricted immunogenicity. Mol Immunol. 1994;31:1391–401. doi:10.1016/0161-5890(94)90155-4.
- Calis JJA, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Kesmir C, Peters B. Properties of MHC Class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9:1–13. doi:10.1371/journal.pcbi.1003266.
- Kardani K, Bolhassani A, Namvar A. An overview of in silico vaccine design against different pathogens and cancer. Expert Rev Vaccines. 2020;19:699–726. doi:10.1080/14760584.2020.1794832.
- Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5:1–14. doi:10.1371/journal.pone.0009862.
- Saha S, Raghava GPS. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct Funct Genet. 2006;65:40–48. doi:10.1002/prot.21078.
- Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi:10.1093/nar/gkx346.
- Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:1–7. doi:10.1186/1471-2105-8-4.
- Dhanda SK, Vaughan K, Schulten V, Grifoni A, Weiskopf D, Sidney J, Peters B, Sette A. Development of a novel clustering tool for linear peptide sequences. Immunology. 2018;155:331–45. doi:10.1111/imm.12984.
- Scholzen A, Richard G, Moise L, Moise L, Baeten LA, Reeves PM, Martin WD, Brauns TA, Boyle CM, Paul SR, et al. Promiscuous Coxiella burnetii CD4 epitope clusters associated with human recall responses are candidates for a novel T-Cell targeted multi-epitope Q fever vaccine. Front Immunol. 2019;10:1–22. doi:10.3389/fimmu.2019.00207.
- Bui HH, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007;8:1–6. doi:10.1186/1471-2105-8-361.
- Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006;7:1–5. doi:10.1186/1471-2105-7-153.
- Kibria KMK, Islam MSB, Ullah H, Miah M. The multi-epitope vaccine prediction to combat Pandemic SARS-CoV-2, an immunoinformatic approach. Research Square Preprint. 2020;00:1–20. doi:10.21203/rs.3.rs-21853/v1.
- Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Kozakov D. New additions to the ClusPro server motivated by CAPRI. Proteins Struct Funct Bioinforma. 2017;85:435–44. doi:10.1002/prot.25219.
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:363–67. doi:10.1093/nar/gki481.
- De Vries SJ, Van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–97. doi:10.1038/nprot.2010.32.
- Kurcinski M, Jamroz M, Blaszczyk M, Kolinski A, Kmiecik S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015;43:419–24. doi:10.1093/nar/gkv456.
- Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–73. doi:10.1093/bioinformatics/btu097.
- Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2:229–34. doi:10.1038/85286.
- Stothard P. The sequence manipulation suite: javaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–04. doi:10.2144/00286ir01.
- Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:526–31. doi:10.1093/nar/gki376.
- Slathia PS, Vaccine DNA. Design for Chikungunya virus based on the conserved epitopes derived from structural protein. Assoc Comput Mach. 2013:849–50. doi:10.1145/2506583.2516950.
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat. Rev Genet. 2008;9:776–88. doi:10.1038/nrg2432.
- Wang S, Zhang C, Zhang L, Li J, Huang Z, Lu S. The relative immunogenicity of DNA vaccines delivered by the intramuscular needle injection, electroporation and gene gun methods. Vaccine. 2008;26:2100–10. doi:10.1016/j.vaccine.2008.02.033.
- Kudela J. Immunization with analog peptide in combination with CpG and montanide expands tumor antigen-specific CD8+ T cells in melanoma patients. J Immunother. 2008;31:781–91. doi:10.1097/CJI.0b013e318183af0b.
- Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, Steinman RM. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med. 2009;206:1589–602. doi:10.1084/jem.20090247.
- Ma J, Wang H, Zheng X, Xue X, Wang B, Wu H, Zhang K, Fan S, Wang T, Li N, et al. CpG/Poly (I:C) mixed adjuvant priming enhances the immunogenicity of a DNA vaccine against eastern equine encephalitis virus in mice. Int Immunopharmacol. 2014;19:74–80. doi:10.1016/j.intimp.2014.01.002.
- Suschak JJ, Williams JA, Schmaljohn CS. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum Vaccines Immunother. 2017;13:2837–48. doi:10.1080/21645515.2017.1330236.
- Henke A, Rohland N, Zell R, Wutzler P. Co-expression of interleukin-2 by a bicistronic plasmid increases the efficacy of DNA immunization to prevent influenza virus infections. Intervirology. 2006;49:249–52. doi:10.1159/000092487.
- Hu H, Tao L, Wang Y, Chen L, Yang J, Wang H. Enhancing immune responses against SARS-cov nucleocapsid DNA vaccine by co-inoculating interleukin-2 expressing vector in mice. Biotechnol Lett. 2009;31:1685–93. doi:10.1007/s10529-009-0061-y.
- Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, Bilska M, Craiu A, Zheng XX, Krivulka GR, et al. Control of viremia prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–92. doi:10.1126/science.290.5491.486.
- Kalams SA, Parker SD, Elizaga M, Metch B, Edupuganti S, Hural J, Rosa SD, Carter DK, Rybczyk K, Frank I, et al. Safety and comparative immunogenicity of an HIV-1 DNA vaccine in combination with plasmid interleukin 12 and impact of intramuscular electroporation for delivery. J Infect Dis. 2013;208:818–29. doi:10.1093/infdis/jit236.
- Jalah R, Patel V, Kulkarni V, Rosati M, Alicea C, Ganneru B, Gegerfelt AV, Huang W, Guan Y, Broderick KE, et al. IL-12 DNA as molecular vaccine adjuvant increases the cytotoxic T cell responses and breadth of humoral immune responses in SIV DNA vaccinated macaques. Hum Vaccines Immunother. 2012;8:1620–29. doi:10.4161/hv.21407.
- Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:1–13. doi:10.3389/fimmu.2019.00594.
- Diken M, Kreiter S, Selmi A, Britten CM, Huber C, Ö T, Sahin U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18:702–08. doi:10.1038/gt.2011.17.
- Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, Shi Y, Sadtler K, Gao W, Lin J, et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol. 2019;37:1174–85. doi:10.1038/s41587-019-0247-3.
- Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–48. doi:10.1038/nrmicro3143.

