ABSTRACT
The emergence of the Omicron SARS-CoV-2 variant of concern has changed the COVID-19 scenario as this variant is characterized by high transmissibility and immune evasion ability. To evaluate the impact of this variant on the Canary Islands (Spain) population, we determined the reinfection rates and disease severity associated with the Omicron sublineages and the previously circulating variants of concern. We performed a retrospective observational study on 21,745 SARS-CoV-2 viral genomes collected from December 2020 to July 2022 in the Canary Islands (Spain). We compared the reinfection rates between lineages using pairwise proportion and Fisher’s exact tests. To assess disease severity, we studied the association of Alpha, Delta, BA.1, BA.2, BA.5, and other risk factors on 28-day hospital mortality using logistic regression and Cox proportional hazard models. We observed 127 bona fide reinfection cases throughout the study period. We found that BA.5 had the highest reinfection rate compared to other lineages (vs. Delta p = 2.89 × 10−25; vs. BA.1 p = 5.17 × 10−11; vs. BA.2 p = 0.002). Among the 1,094 hospitalized patients, multivariate logistic regression showed that Alpha (Odds Ratio [OR] = 0.45, 95% Confidence Interval [CI] = 0.23-0.87, p = 0.02), BA.2 (OR = 0.38, 95% CI = 0.22-0.63, p = 1.91 × 10−4), and BA.5 (OR = 0.30, 95% CI = 0.16-0.55, p = 1.05 × 10−4) had lower 28-day hospital mortality compared to Delta. These results were confirmed by using Cox proportional hazard models. Omicron lineages, and in particular BA.5, were associated with higher reinfection rates and lower disease severity (28-day hospital mortality) than previously circulating variants of concern.
Introduction
The Omicron (B.1.1.529) SARS-CoV-2 variant of concern (VOC) has been responsible for the sixth coronavirus disease of 2019 (COVID-19) wave at the beginning of 2022 in Europe and worldwide. Studying clinical severity and reinfection rates associated with this VOC is important to understand the public health impact and provide clinical guidance, especially in a scenario where novel Omicron-derived sublineages have emerged, e.g. BA.2 or BA.5. Previous studies have shown that BA.1 and BA.2 lineages are associated with lower disease severity compared to the previously circulating Delta variant [Citation1], despite being responsible for a surge of infections. Cases of reinfection have also increased after December 2021, corresponding to the emergence of the Omicron variant in many countries [Citation2,Citation3]. More recent studies have shown that BA.4/BA.5 infections are characterized by increased immune evasion compared to BA.1/BA.2 and Delta (B.1.617.2 and sublineages), despite no differences in disease severity have been found between these lineages [Citation4]. We have previously shown that Delta and its sublineages were associated with higher 28-day mortality than Alpha (B.1.1.7) in hospitalized patients from the Canary Islands region (Spain) [Citation5,Citation6]. By expanding the dataset of viral genome sequences obtained in the region, here we first compared the reinfection rates of circulating variants in a period from December 2020 to July 2022. Then, we evaluated disease severity by assessing the association of the infections with Alpha, Delta, BA.1, BA.2, and BA.5 and 28-day mortality among hospitalized COVID-19 patients.
Materials and methods
Sample collection, sequencing, and lineage assignment
The study was conducted at the University Hospital Nuestra Señora de Candelaria (HUNSC) (Santa Cruz de Tenerife, Spain). The institutional review board approved the study (approval number: CHUNSC_2020_24).
Nasopharyngeal swab samples were collected using standard procedures and RT-qPCR reactions were performed to assess SARS-CoV-2 positivity, as previously described [Citation7]. Positive samples with a Ct ≤30 were included for sequencing.
Libraries were prepared using two alternative tiling amplicon sequencing protocols. Illumina, Inc. sequencing was conducted using the COVIDSeq test based on ARTIC V3 (from the 18th December 2020 to the 15th April 2022) or V4 (from the 15th April to the 26th July 2022) and a NextSeq 550 instrument on High Output mode with 36 paired-end reads. Oxford Nanopore Technologies (ONT) sequencing was conducted using the COVID Mini kit (ONT), a MinION sequencing device, and R9.4 flow cells. MinKNOW software and the embedded Guppy software (v.5) were used for sequencing tuning and basecalling. DRAGEN (v1.3.0.28) COVIDSeq test and viralrecon (v2.5 or previous versions) [Citation8] were used for read filtering and consensus generation for Illumina and ONT data, respectively. Lineages were assigned using Pangolin v.4.1.2 in the Usher mode [Citation9], while sequence quality control (QC) was carried out using Nextclade (v2.3.1) [Citation10]. Only those sequences that had an assigned lineage and a percentage of Ns < 10% were categorized as having a “good” or “median” QC and were used for the analysis. Sequences collected up to the 27th of September 2021 are part of previous studies [Citation5,Citation6].
Patient inclusion and exclusion criteria for studies
Reinfection rates were calculated by normalizing the number of reinfections caused by one lineage by the total number of samples assigned to that lineage in our dataset. To focus on bona fide reinfections, we defined as reinfected those individuals that had two sequenced positive SARS-CoV-2 samples with a time distance of >90 days. Following guidelines, we also included as reinfections those cases with different lineages in primary and secondary infections, defining as different lineages those that have sequences with more than two-nucleotide difference per each month separating the sample collections [Citation11]. Within these, we included only those samples with a time distance between sample collection of >30 days. To exclude the possibility of persistent infections in patients with more than two sequenced samples collected throughout the study period, we included only those with different lineages and with a collection time distance of >60 days.
The study of the association of variants with disease severity was conducted on patients hospitalized at the Complejo Hospitalario Nuestra Señora de Candelaria and the Hospital Universitario de Canarias (Tenerife island, n = 1,079), Complejo Hospitalario Universitario Insular Materno Infantil (Gran Canaria island, n = 28), Hospital General de Fuerteventura (Fuerteventura island, n = 8), Hospital Universitario Doctor José Molina Orosa (Lanzarote island, n = 8), Hospital General de La Palma (La Palma island, n = 19), Hospital Nuestra Señora de Los Reyes (El Hierro island, n = 2). We excluded from the study those patients admitted to the hospital for reasons other than COVID-19 or that did not have symptoms related to COVID-19. We defined as hospitalized only those patients that were hospitalized between 0 and 21 days from the onset of symptoms, and deceased only those patients that died within 28 days from hospital admission. The patients that did not have a reported discharge date were excluded from the analysis.
Statistical analysis
Descriptive statistics were reported as frequencies with percentage and included patient age, sex, comorbidities, length of hospital stay, ICU admission, and mortality. To evaluate whether there was a difference in reinfection rates between lineages, we performed a pairwise proportions test with post hoc Holm correction or a Fisher’s exact test with post hoc Holm correction. To assess the association between the lineage causing the infection and 28-day hospital mortality, we used logistic regressions and Cox proportional hazards models, including lineage, age, sex, length of hospital stay, and the presence of one or multiple comorbidities as covariates. Statistical analysis was performed in R software (v3.6.3) [Citation12]. Statistical significance was set at p < 0.05.
Results
Reinfection rates increased with BA.2 and BA.5 lineages
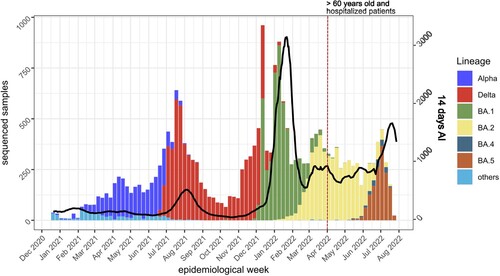
During the period from the 19th of December 2020 to the 26th of July 2022, we sequenced 27,363 samples from COVID-19 patients in the Canary Islands region. Of these, 21,745 samples passed the QC and obtained an assigned lineage. shows the distribution of samples by variant and the 14-day accumulated incidence (AI) of COVID-19 cases in the Canary Islands. A total of 2,859 samples were assigned to Alpha (B.1.1.7 and sublineages), 6,637 samples to Delta (B.1.617.2 and sublineages), 4,977 to BA.1, 5,013 to BA.2, 173 to BA.4, and 1,430 to BA.5. We found recombinant lineages in 14 samples (seven XE and seven XU lineages).
Figure 1. 14-day accumulated incidence (AI) and variants observed in the Canary Islands from December 2020 to July 2022. Note that, from the end of March 2022 (red dotted vertical line), only samples from >60 years old or hospitalized patients were sequenced and included in the 14-days AI calculation by public health authorities. Black line, 14-days AI.
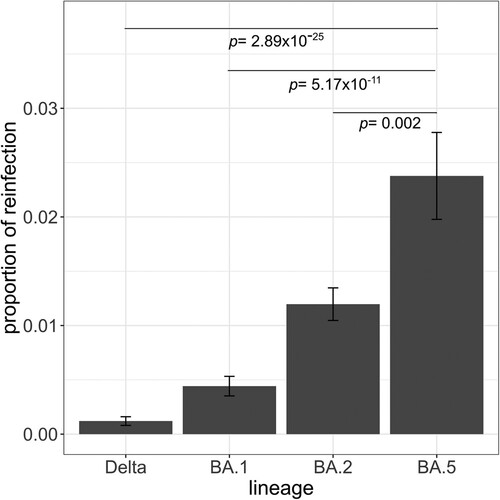
Throughout the study period and only within the sequenced samples, we observed a total of 127 cases of bona fide reinfections, of which two were of patients that were reinfected twice. We found that BA.5 was the variant associated with the highest proportion of reinfections throughout the study period, as more than 2% of the cases associated with this variant were reinfection cases (pairwise proportions test with post hoc Holm correction, vs. Delta p = 2.89 × 10−25; vs. BA.1 p = 5.17 × 10−11; vs. BA.2 p = 0.002) (). We excluded BA.4 from this analysis because of the low number of reinfection cases assigned to this variant (n = 4). In our dataset, we did not observe any reinfected individual with variants circulating before Delta.
Figure 2. Proportion of reinfections observed in the Canary Islands by lineage. The proportion of reinfections was calculated as the number of reinfections by lineage divided by the total number of infections by that lineage. Pairwise proportions tests with post hoc Holm correction were applied. Only p values between BA.5 and the other lineages are shown. Error bars represent the standard error of sample proportion.
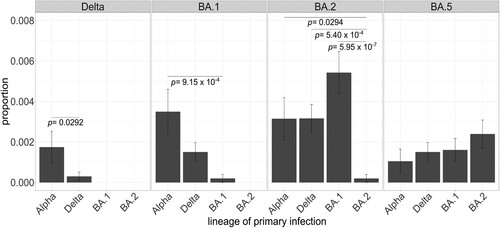
Within these samples, there were four cases of reinfection with the same variants (including two with different sublineages of Delta, and two with different sublineages of BA.1 and BA.2). For Delta and BA.1, the number of reinfections was higher for lineages that emerged earlier in the pandemic (e.g. Alpha) than those that dispersed more closely to the lineage causing the reinfection (Delta). In contrast, this pattern was not observed for BA.2 and BA.5 since both showed similar reinfection rates for all lineages (). Not surprisingly, the reinfection rate for BA.2 was significantly lower when primary infection was caused by another BA.2 sublineage than by other lineages.
| 2. | Omicron lineages are associated with reduced risk of disease severity among hospitalized patients compared to Delta | ||||
Figure 3. Proportion of primary infections in lineage-causing reinfections. Each panel encompasses the lineage causing the secondary infection and each bar represents the number of reinfection cases by lineage causing the primary infection normalized by the total number of cases assigned to that lineage throughout the study period. Fisher’s exact tests with post hoc Holm correction were applied to test the significance of the intra-group differences. Only significantly different comparisons are shown. Error bars represent the standard error of sample proportion. Reinfection cases with primary infection with n = 1 were excluded.
A total of 1,144 patients were hospitalized throughout the study period and met the inclusion criteria (). summarizes the demographic and clinical characteristics of the patient population, as a whole and stratified by VOC lineage. We observed a broader distribution of age for patients infected with Alpha and Delta lineages compared to those with the Omicron sublineages, who had a distribution shifted towards older ages (Table 1, Figure S1). The median age of hospitalized patients infected with Alpha or Delta was lower than those infected with Omicron (Alpha = 57; Delta = 59; BA.1 = 73; BA.2 = 80; and BA.5 = 80.5 years old).
Figure 4. Maximum likelihood phylogenetic tree of SARS-CoV-2 sequences (radial) placing the sequences from 1,144 hospitalized cases with infections in the Canary Islands throughout the study period. The tree also includes relationships with other 1,504 genome sequences representative of major clades (light grey), labelled based on Nextstrain definitions, from Nextclade v2.13.0 and Auspice v2.24.2 (as of 2023-03-24), and sampled between December 2019 and July 2022 for reference.
Table 1. Demographic and clinical data of hospitalized cases with Alpha, Delta, BA.1, BA.2, and BA.5 infections in the study period.
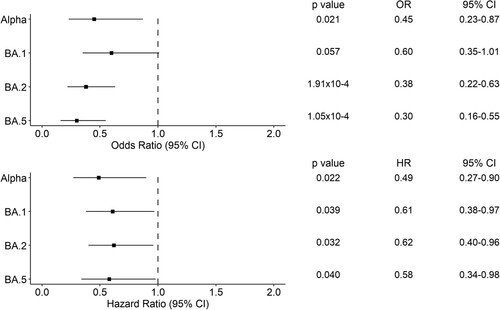
To assess whether Omicron lineages were associated with reduced risk to disease severity than Delta, we performed a logistic regression analysis using patient 28-day hospital mortality as the dependent variable. We found that Alpha (Odds Ratio [OR] = 0.45, 95% Confidence Interval [CI] = 0.23-0.47, p = 0.021), BA.2 (OR = 0.37, 95% CI = 0.22-0.63, p = 1.91 × 10−4) and BA.5 (OR = 0.30, 95% CI = 0.16-0.55, p = 1.05 × 10−4) were associated with lower 28-day hospital mortality than Delta (, Table S1). Cox proportional hazards model also provided the same findings (Alpha, Hazard Ratio [HR] = 0.49, 95% CI = 0.27-0.90, p = 0.022; BA.2, HR = 0.62, 95% CI = 0.40-0.96, p = 0.032; BA.5, HR = 0.58, 95% CI = 0.34-0.98, p = 0.040), where also BA.1 was found having significantly lower 28-day hospital mortality than Delta (HR = 0.61, 95% CI = 0.38-0.97, p = 0.039). Higher 28-day hospital mortality was also associated with being older (p = 3.63 × 10−10), being male (p = 0.005), and the presence of comorbidities (p = 0.014) (Table S1).
Figure 5. Forest plots of the 28-day hospital mortality associated with the presence of a variant of concern compared to Delta. The upper panel shows the estimated odds ratio (OR) and the 95% confidence interval (CI) from the logistic regression analysis. The lower panel shows the hazard ratio (HR) and the 95% CI from the Cox regression analysis. P-values are also reported.
Discussion
In this study, we investigated the reinfection rate and disease severity of the SARS-CoV-2 VOCs circulating in the Canary Islands (Spain) throughout the COVID-19 pandemic starting from mid-December 2020. First, we compared the reinfection rates of Delta, BA.1, BA.2, and BA.5 in COVID-19 patients from the region since we were unable to unequivocally identify reinfection cases assigned to previously circulating lineages. We found that BA.5 was associated with the largest proportion of reinfection cases in our study, with most of the cases having a primary infection assigned to another Omicron variant (BA.1 or BA.2) which was circulating in the region only two to six months earlier. We observed that for Delta and BA.1 most primary infections were assigned to variants circulating earlier in the pandemic (e.g. Alpha), pointing to an immunity protection after a more recent infection. However, this scenario was not observed for the other VOCs. In fact, the distribution of primary infections for BA.2 and BA.5 was more homogeneous among the lineages. These observations suggest that the higher reinfection rates of BA.2 and BA.5 could be mostly due to immune evasions from prior infections rather than population waning immunity, as previously suggested [Citation2]. In line with our observations, BA.2 and, to a larger extent, BA.4/BA.5 display increased evasion capacity from neutralizing antibodies compared to previously circulating variants [Citation13–15]. Furthermore, BA.4/BA.5 have been shown to confer increased resistance to monoclonal antibodies, a characteristic that has been mainly assigned to the presence of the L452R and the F486V mutations in the Spike gene of these variants [Citation16].
We also evaluated the association of infections by the different VOCs with 28-day hospital mortality. We first assessed whether there was a difference in the hospitalized patient population age in the lineage groups. We observed a wider patient age distribution towards younger age ranges for Alpha and Delta lineages than for Omicron lineages, with the median age higher for the latter. Changes in the non-pharmacological interventions by public health authorities in the population (following only patients 60 years old or older from the end of March 2022) can hardly explain this observation. Instead, we assigned this partly to the different vaccination status of patients throughout the pandemic waves. Besides, the hospitalized patients for Omicron could be older compared to those with Delta or Alpha infections simply because the patients had to accumulate comorbidities to trigger severe symptoms. In fact, during the Alpha surge, the majority of the population had not received the first vaccine dose yet (only 1-35% of the population with ≥1 dose between January and June 2021), while a higher percentage of individuals had received the two vaccine doses during the Delta wave (50-70% of the population with two doses between July and September 2021). On the other hand, during the Omicron wave, most of the Canary Islands inhabitants had received complete vaccination (76%−80% of the population with full vaccination) and many had already received the booster dose (https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/pbiVacunacion.htm). Furthermore, during the Omicron surges, younger adults acquired a higher level of natural immunity after transmission of previous variants, which may have affected the hospitalized patient population.
In terms of disease severity, besides confirming previously published results on Alpha [Citation5], our study supports that BA.2 and BA.5 were associated with lower risk for 28-day hospital mortality than Delta. This confirms previous studies showing that Omicron lineages have reduced disease severity compared to Delta [Citation1]. Furthermore, we did not find any difference in disease severity between BA.5 lineage and previously circulating Omicron sublineages, consistent with results from studies conducted in patients from Southern California [Citation4] and South Africa [Citation17].
This study has several limitations. First, in the reinfection study, our dataset is subjected to the ascertainment bias since we only included in the analysis those samples that were sequenced, excluding those individuals that have had a primary and secondary infection but only one of their samples was sequenced. Second, we were unable to analyse the same proportion of samples out of the total number of incident cases throughout the pandemic, with this proportion being lower during the Omicron surge when we observed the highest incidence of COVID-19 cases in the region. An intrinsic limitation of the study may be due also to the lower viral loads usually found in the nasopharyngeal swabs of patients infected with some of the Omicron variants (e.g. BA.1 [Citation18] or BA.5 [Citation19]) compared to those with other variants. By limiting the sequencing of those positive samples with Ct ≤30, some patients infected with these Omicron variants may have been missed, possibly biasing the results. Furthermore, because of the relaxation of the COVID-19 control measures in Spain, only samples obtained from patients older than 60 or those hospitalized of any age were collected for viral genome sequencing in the Canary Islands from the end of March 2022. Another limitation that needs to be pointed out, especially for the study of viral variant associations with disease severity, is that we had no access to patient vaccination status, and therefore we could not control the association models for this variable. To limit the impact of the vaccination status on the results, as we did previously [Citation5], we restricted our analysis to hospitalized patients only.
Supplemental Material
Download MS Word (174.9 KB)Acknowledgements
We would like to deeply thank the patients participating in the study. JMLS, HRP, and AMB acknowledge the University of La Laguna for the training support during the PhD studies.
Disclosure statement
No potential conflicts of interest were reported by the authors.
Data availability statement
Sequences in FASTA format and associated metadata have been deposited in GISAID (https://gisaid.org) under identifiers EPI_SET_230213uw and EPI_SET_230322qh.
Additional information
Funding
References
- Sievers C, Zacher B, Ullrich A, et al. SARS-CoV-2 Omicron variants BA.1 and BA.2 both show similarly reduced disease severity of COVID-19 compared to Delta, Germany, 2021 to 2022. Eurosurveillance. 2022: 27.
- Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:eabn4947.
- Ruff J, Zhang Y, Kappel M, et al. Rapid increase in suspected SARS-CoV-2 reinfections, Clark County, Nevada, USA, December 2021. Emerg Infect Dis J. 2022: 28.
- Lewnard JA, Hong V, Tartof SY. Association of SARS-CoV-2 BA.4/BA.5 Omicron lineages with immune escape and clinical outcome. medRxiv. 2022;2022.07.31.22278258. Available from: http://medrxiv.org/content/early/2022/08/02/2022.07.31.22278258.abstract.
- Ciuffreda L, Alcoba-Florez J, Lorenzo-Salazar JM, et al. Association of the Delta SARS-CoV-2 variant with 28-day hospital mortality between December 2020 and September 2021. J Infect. 2022;85:90–122.
- Ciuffreda L, González-Montelongo R, Alcoba-Florez J, et al. Tracing the trajectories of SARS-CoV-2 variants of concern between December 2020 and September 2021 in the Canary Islands (Spain). Front Cell Infect Microbiol. 2022;12:919346.
- Alcoba-Florez J, Gil-Campesino H, Artola D-M de, et al. Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection. Int J Infect Dis. 2020;99:190–192.
- Patel H, Varona S, Monzón S, et al. nf-core/viralrecon: nf-core/viralrecon v2.5. 2022.
- O’Toole Á, Scher E, Underwood A, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064.
- Aksamentov I, Roemer C, Hodcroft EB, et al. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6:3773.
- World Health Organization. Public health surveillance for COVID-19: interim guidance. Geneva, Switzerland; 2022. Available from: https://www.who.int/publications/i/item/WHO-2019-nCoV-SurveillanceGuidance-2022.2.
- R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2020; Available from: https://www.r-project.org/.
- Cao Y, Yisimayi A, Jian F, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602.
- Hachmann NP, Miller J, Collier AY, et al. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. 2022;387:86–88.
- Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13.
- Wang Q, Guo Y, Iketani S, et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608.
- Wolter N, Jassat W, Walaza S, et al. Clinical severity of SARS-CoV-2 Omicron BA.4 and BA.5 lineages compared to BA.1 and Delta in South Africa. Nat Commun. 2022;13:5860.
- Sentis C, Billaud G, Bal A, et al. SARS-CoV-2 omicron variant, lineage BA.1, is associated with lower viral load in nasopharyngeal samples compared to delta variant. Viruses. 2022: 14.
- Takatsuki Y, Takahashi Y, Nakajima J, et al. Viral load of SARS-CoV-2 Omicron BA.5 is lower than that of BA.2 despite the higher infectivity of BA.5. Immunity, Inflamm Dis. 2023;11:e783.
- Rees EM, Nightingale ES, Jafari Y, et al. COVID-19 length of hospital stay: a systematic review and data synthesis. BMC Med. 2020;18:270.

