?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Between January 2015 and October 2022, 38 patients with culture-confirmed melioidosis were identified in the Kowloon West (KW) Region, Hong Kong. Notably, 30 of them were clustered in the Sham Shui Po (SSP) district, which covers an estimated area of 2.5 km2. Between August and October 2022, 18 patients were identified in this district after heavy rainfall and typhoons. The sudden upsurge in cases prompted an environmental investigation, which involved collecting 20 air samples and 72 soil samples from residential areas near the patients. A viable isolate of Burkholderia pseudomallei was obtained from an air sample collected at a building site five days after a typhoon. B. pseudomallei DNA was also detected in 21 soil samples collected from the building site and adjacent gardening areas using full-length 16S rRNA gene sequencing, suggesting that B. psuedomallei is widely distributed in the soil environment surrounding the district. Core genome-multilocus sequence typing showed that the air sample isolate was phylogenetically clustered with the outbreak isolates in KW Region. Multispectral satellite imagery revealed a continuous reduction in vegetation region in SSP district by 162,255 m2 from 2016 to 2022, supporting the hypothesis of inhalation of aerosols from the contaminated soil as the transmission route of melioidosis during extreme weather events. This is because the bacteria in unvegetated soil are more easily spread by winds. In consistent with inhalational melioidosis, 24 (63.2%) patients had pneumonia. Clinicians should be aware of melioidosis during typhoon season and initiate appropriate investigation and treatment for patients with compatible symptoms.
Introduction
Melioidosis is caused by the aerobic Gram-negative bacillus Burkholderia pseudomallei, which is ubiquitous throughout the tropics, particularly in Southeast Asia and northern Australia [Citation1,Citation2]. Hong Kong is also considered to be within the risk zone due to the high environmental suitability for B. pseudomallei [Citation1,Citation2]. B. pseudomallei is a natural saprophyte [Citation3], and percutaneous inoculation, ingestion, and inhalation of contaminated soil or water are well-recognized modes of transmission of melioidosis[Citation4]. The disease was first recognized in Burma in 1911 [Citation5]. The earliest report of melioidosis in Hong Kong dates back to the outbreak in marine mammals at an oceanarium in 1975 and 1976 [Citation6], with the first case series of human melioidosis in Hong Kong reported in 1984, which documented five local cases of septicemia [Citation6]. Over the last two decades, an increasing trend of 61 cases has been identified, indicating that melioidosis has become endemic in Hong Kong [Citation7].
Melioidosis patients present a wide clinical spectrum ranging from localized infections, pneumonia, to multiple-organ involvement and sepsis [Citation8]. Melioidosis often affects individuals with preexisting conditions associated with an altered immune response, with diabetes mellitus, hazardous alcohol use, chronic lung disease, and chronic kidney disease being the most common risk factors identified [Citation8]. The increased susceptibility may be related to polymorphonuclear neutrophil defects [Citation9], and such conditions may be critical in determining whether an infection is established after exposure to B. pseudomallei [Citation10]. The incubation period varies, usually from 2 to 4 weeks, but can be shorter with a high inoculum [Citation11]. In some cases, melioidosis may remain latent for months or years before symptoms develop; the longest documented latency between exposure and clinical melioidosis is 62 years [Citation12].
There is a strong association between melioidosis incidence and rainfall intensity [Citation13,Citation14]. Epidemiological studies have supported inhalation as the mode of transmission after heavy rains and winds, although there is limited research regarding the sampling of ambient B. pseudomallei for phylogenetic confirmation [Citation15,Citation16]. Between January 2015 and October 2022, 38 patients with the first episode of culture-confirmed melioidosis were identified in the Kowloon West (KW) Region of Hong Kong, with 30 of them clustered in the Sham Shui Po (SSP) district within an estimated area of 2.5 km2. Notably, 18 patients were identified from August to October 2022 after heavy rainfall and typhoons. The sudden increase in cases in conjunction with the highly localized, urban place of residence of the patients was unusual and prompted further epidemiological investigations. The aim of the study was to determine the transmission dynamics of this urban outbreak of melioidosis. Bacterial culture and 16S rRNA gene-based metagenome sequencing were used to determine the presence and distribution of B. pseudomallei in the environment, including ambient air and soil. Core genome multilocus sequence typing (cg-MLST) was applied to study the molecular epidemiology of the clinical isolates collected from 2015 to 2022 in KW region and to demonstrate their phylogenetic relatedness with the environmental isolate. A quantitative analysis of multispectral satellite images was conducted to study the change in vegetation region in the SSP district from 2015 to 2022, which determined the soil integrity and the likelihood of aerosolization of bacteria in soil under strong winds. The study protocol was approved by the Kowloon West Cluster Clinical Research Ethics Committee, Hospital Authority [KW/EX-22-077(176-05)].
Materials and methods
Clinical setting
The KW region is served by four regional hospitals with a total of 3,431 beds, catering to an estimated population of 1.4 million. Cases of the first episode of melioidosis infection from January 1, 2015, through October 31, 2022, were identified based on bacterial culture records of the region-based microbiological laboratory.
Clinical data retrieved from medical records included patient demographics, comorbidities, duration of symptoms, type of infection, length of hospital stay, intensive-unit care, and antimicrobial therapy. Immunosuppression was defined as the presence of haematologic malignancy, solid organ transplantation, diabetes mellitus, chronic renal failure, alcoholism, cirrhosis, human immunodeficiency virus (HIV) infection, history of corticosteroid use, chemotherapy, or monoclonal antibody use [Citation17]. Laboratory data, namely leukocyte counts and C-reactive protein levels, were also included. The major outcome measures were 7-day and 30-day mortality.
Environmental sampling
Prospective collection of air and soil samples was conducted near the residence of affected patients in the SSP district from August to October 2022. For air collection, 1000 L of air was passed (180 L/min for 5.5 min) through a portable microbiologic air sampler (VWR SAS Super ISO 180) directly on an Ashdown’s agar consisting of trypticase soy agar with 4% glycerol, 5 ml/L crystal violet, 50 mg/L neutral red, and 4 mg/L gentamicin. After air sampling, the agar was incubated at 35°C aerobically for 10 days.
Soil samples were collected and cultured as previously described [Citation15]. In brief, soil samples were collected at approximately 10 cm below the surface. Sterile water (20 mL) was added to 20 g of soil and shaken at 220 rpm for 48 h at 35°C. After the soil samples were removed from the shaker and left to stand for 1-2 h, 10 mL supernatant was placed in 30 mL of modified Ashdown selective broth, consisting of 10 g/L tryptone soy broth, 40 ml/L glycerol, 5 mg/L crystal violet and 50 mg/L colistin, and incubated at 35°C. The broth supernatant was plated on Ashdown’s agar after 2 and 7 days, and the cultures were incubated aerobically at 35°C for 10 days.
Identification and genotyping of B. pseudomallei
The colonies grown on agar plates were screened for the morphotype of B. pseudomallei. Suspected colonies were initially identified as Burkholderia thailandensis using matrix-assisted laser desorption/ionization time-of-flight (MALDI Biotyper, Bruker Daltonics; Database version: IVD-9468 MSP). Real-time PCR targeting the Type III secretion system of B. pseudomallei was used to confirm all isolates from the suspected colonies [Citation18]. All first positive isolates were tested for susceptibility against meropenem, imipenem, ceftazidime, amoxicillin/clavulanate, doxycycline, tetracycline and trimethoprim/sulfamethoxazole using Etest (Liofilchem ®, Italy) following the manufacturer’s instructions. The minimum inhibitory concentrations (MICs) were interpreted according to the breakpoints of Clinical and Laboratory Standards Institute (CLSI) M45-A3 [Citation19].
Whole-genome sequencing of B. pseudomallei isolates
Total nucleic acid of B. pseudomallei isolates was extracted using the QIAamp BiOstic Bacteremia DNA Kit (Qiagen, Germany) as per the manufacturer’s protocol. The library for Nanopore sequencing was prepared using the Rapid Barcoding Kit 96 (SQK-RBK110.96) following the manufacturer’s protocol. The normalized library (800 ng) was loaded on the R9.4.1 flow cell (FLO-MIN106) and sequenced using the GridION MK1 sequencing device (ONT, UK). Basecalling was performed using Guppy v6.2.7 with super-accurate basecalling (SUP) mode. The filtered reads were analyzed using dragonfly v1.0.12, an automated pipeline for de novo assembly of bacterial genomes. Adapter sequences were removed using Porechop v0.2.4, and sequences with lengths below 1000 bases were filtered out using Nanoq v.0.9.0. The genome size was estimated to be 7.1 Mbps, and reads were randomly subsampled to 100X coverage using Rasusa v0.6.1. De novo assembly was performed using Flye v2.9-b1768. The draft genomes were polished by one round of Racon v1.5.0 and one round of Medaka v1.5.0. Contigs less than 500 bases were discarded.
The raw reads and closed-gap complete genomes of 38 isolates of B. pseudomallei have been submitted to the NCBI GenBank under the BioProject no. PRJNA904945.
Core-genome multilocus sequence typing (cgMLST)
A total of 150 genomes of B. pseudomallei, annotated as “Chromosome” or “Complete”, and containing exactly two chromosome sequences were extracted from the NCBI assembly database. Multilocus sequence typing (MLST) was used to assign a sequence type (ST) to each isolate. The sequences of the seven MLST alleles (ace, gltB, gmhD, lepA, lipA, nark, ndh) were extracted from the B. pseudomallei genome sequences using NCBI Blastn search (sequence identity ≥ 99.5% and subject coverage = 100%) [Citation20,Citation21]. The ST was assigned based on the MLST allele profile accessible on the Public Databases for Molecular Typing and Microbial Genome Diversity (PubMLST) [Citation22]. Core genome MLST (cgMLST) genes of B. pseudomallei (n = 4090) were downloaded from the PubMLST [Citation22]. Gene coding sequences were predicted from the assembled whole genome contigs of the B. pseudomallei collected in this study and the reference B. pseudomallei genomes using PROKKA [Citation23]. The gene coding sequences were aligned to the B. pseudomallei cgMLST reference genes using Diamond Blastx [Citation24]. Alignments with sequence identity ≥ 90% and subject coverage ≥ 95% to the reference genes were defined as encoding a cgMLST gene (modified from Ref. [Citation22]). Single-copy cgMLST genes that exist in all B. pseudomallei genomes were used in the subsequent analyses to compare Hong Kong and global B. pseudomallei isolates. Codon-based multiple sequence alignment was performed using MAFFT [Citation25] on each single copy cgMLST gene. All single copy cgMLST gene alignments were concatenated into a single nucleotide sequence alignment. Sites with single nucleotide polymorphism (SNP) were used to estimate the maximum likelihood phylogeny using GTR+Γ nucleotide substitution model in RAxML [Citation26]. Phylogenetic robustness was evaluated using bootstrap analysis of 1000 pseudoreplicates of the sequences. The resulting tree was visualized with the metadata (source of specimen, country and year of collection, if available) using the ggtree package in R-3.6 [Citation27]. The codes for the cgMLST analysis and tree visualization were available in Github repository https://github.com/id-bioinfo/Publication-Materials/tree/main/B.Pseudomallei_2022.
In silico pulsed-field gel electrophoresis (PFGE)
Thirty-eight isolates with complete circular chromosome 1 and chromosome 2, and 12 related reference genomes were selected for in silico PFGE. The genomic position of the AflII restriction enzyme cleavage was searched in the two chromosomes of each B. pseudomallei isolate, and the fragment size was calculated based on the predicted cleavage positions. A UPGMA dendrogram was generated using GelJ [Citation28] to show the clustering of the fragment size profiles based on Dice dissimilarity. The genetic relatedness among the isolates was interpreted according to the criteria described by Tenover et al. [Citation29].
16s rRNA gene sequencing for soil samples
To extract total nucleic acid from 1 mL of soil supernatant, homogenization with purified water was performed, followed by extraction using the QIAamp BiOstic Bacteremia DNA Kit (Qiagen). The full length of 16S rRNA gene of each sample was amplified and barcoded using the 16S Barcoding Kit 1-24 (SQK-16S024) from ONT according to the manufacturer’s protocol. Amplicons were purified with AMPure XP beads (Beckman Coulter) and quantified using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) with Qubit™ 1X dsDNA HS Assay Kit (Thermo Fisher Scientific). A maximum of 24 barcoded libraries were pooled into one tube in equal concentrations. The pooled library was then loaded onto the flow cell FLO-MIN106 R9.4.1 and sequenced for approximately 24 h using the GridION sequencer. Basecalling was performed on the sequencing platform MinKNOW using the “super accuracy” mode. During sequencing, the fastq files with a quality score ≥ 10 were simultaneously uploaded onto the cloud-based analysis platform Epi2me for real-time analysis. The FASTQ 16S workflow (v2022.01.07) on Epi2me was used to classify the 16S rRNA genes in the samples. The parameter settings of the workflow were: QSCORE ≥ 10, minimum length filter 1000, maximum length filter 2000, minimum percentage coverage 90% and minimum percentage identity 90%.
Vegetation region of SSP retrieval from satellite imagery
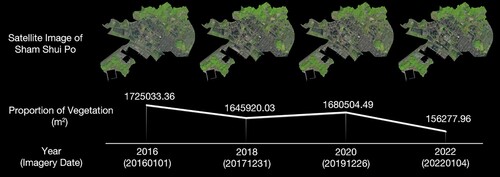
The vegetation region of SSP was determined for the years 2016, 2018, 2020, and 2022 using Sentinel-2 satellite imagery with a 10-meter resolution. These multispectral images were obtained from the year 2015 and cover the Hong Kong region. The satellite data were obtained from USGS (U.S. Geological Survey). In order to ensure the vegetation in each image was captured during the same season, images were selected as close to January 1st each year and without cloud cover. Consequently, the four images were dated January 1, 2016, December 31, 2017, December 26, 2019, and January 4, 2022. The Normalized Difference Vegetation Index (NDVI) was used to quantify vegetation by measuring the difference between near-infrared (which vegetation strongly reflects) and red light (which vegetation absorbs). It is calculated by the formula (1) below:
(1)
(1) where NIR is the near-infrared band value of the imagery and RED is the visible-red band value of the imagery. This allowed vegetation to be distinguished from other land covers through image classification. Following supervised image classification, the vegetation region was calculated.
Wind retrieval
Inverse Distance Weighted (IDW) interpolation algorithm was applied to simulate the wind data in the target area. The formula for IDW is
(2)
(2) where
is the interpolation result of point
,
is the observation value of point
, and
is the
th power of the distance from the point
to point
. The IDW algorithm utilized the principle of the first law of geography, which states that closer objects have a higher similarity in attributes and thus should carry more weight [Citation30]. Specifically, the weight assigned by the IDW algorithm decreased as the distance from the sampled location increased [Citation31]. This method has been widely employed for wind retrieval at the city level [Citation32–34]. In this study, wind data was simulated using IDW based on data collected from 8 Hong Kong Observatory stations surrounding SSP. The station closest to the air sampling site was King's Park, which was approximately 2.8 km away. Wind direction and speed data from the other 7 stations, namely Kai Tak, Sha Tin, Star Ferry, Tai Po Kau, Tate's Cairn, and Tsing Yi, were used to estimate the wind data of the target area during the collection of the positive air sample on August 15, 2022, from 10:00–11:00 am.
Statistical analysis
A Poisson regression model was employed to investigate the statistical correlation between melioidosis cases and adverse weather events. To quantify the effects of typhoons effects, a weighted typhoon signal scoring system was utilized, which was modified from a previous study [Citation35]. Under this system, Typhoon signal No.1–3 was assigned to 1 point, Typhoon signal No.8 received 3 points, and Typhoon signal No.9–10 was assigned 5 points. The odds of melioidosis cases caused by prior exposure to rainfall and typhoons were computed using RStudio (version 2022.12.0 Build 353) and the Poisson regression model. Statistical significance was determined by a p-value of less than 0.05 for all statistical tests.
Results
Demographic and clinical characteristics of melioidosis in the KW region
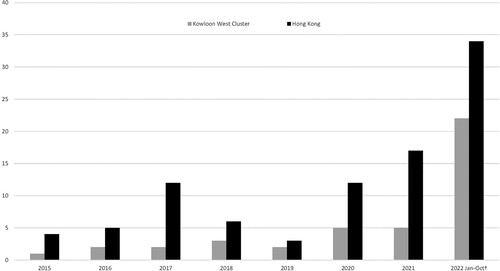
A total of 38 patients with culture-proven melioidosis, who had their first episode between January 1, 2015, and October 31, 2022, were identified. Among them, 29 patients were admitted to Caritas Medical Centre, seven were admitted to Princess Margaret Hospital, and two were admitted to Yan Chai Hospital in the KW Region. The KW Region, which corresponds to a 19% population catchment, was overrepresented, accounting for 46% of the local melioidosis cases, and an increasing trend has been observed since 2019 ().
Figure 1. Number of culture-confirmed melioidosis# in Hong Kong from Jan 2015 to Oct 2022. Note: #Data is retrieved from the Clinical Data Analysis and Reporting System (CDARS), Hospital Authority, Hong Kong. Each patient is counted once per year based on the first detection of the year. †From January to July 2022, 4 cases of melioidosis were diagnosed; from August to October 2022, an unusual upsurge of 18 cases of melioidosis was detected in the Kowloon West Cluster.
The median age of the patients was 68.5 years, ranging from 42 to 94 years, and 32 (84.2%) of them were male (). A total of 27 (71.1%) patients had immunosuppression, with diabetes mellitus (50%) being the most common comorbidity. Most patients presented with pneumonia (63.2%), and 78.9% of patients had bacteremia. Six patients (15.8%) had multiple sites of infection. The overall case fatality rate was 26.3%.
Table 1. Characteristics of patients with melioidosis and Burkholderia pseudomallei isolates in Kowloon West Region, Hong Kong, China, January 2015 to October 2022.
Three isolates (7.9%, 3/38) were resistant to trimethoprim/sulfamethoxazole, while all other isolates were susceptible to other antibiotics tested ().
In silico PFGE and phylogenetic analysis of local melioidosis
Similarity search among the global B. pseudomallei reference genomes and assembled genomes from the Hong Kong isolates collected in this study has identified 1,652 single-copy cgMLST genes present in all genomes. A phylogeny constructed using the SNPs of these cgMLST genes revealed that 36 out of 38 KW isolates formed three well-supported (bootstrap = 100%) monophyletic clades (grey, blue and orange blocks in ). The larger clade (grey) consisted of 30 B. pseudomallei isolates closest to the previously reported Isolate 982 from Malaysia in 2015 with a different ST (). The isolates had 96.45% to 97.90% SNP identities in the 1,652 cgMLST genes, and 89.7% (26/30) of these isolates were collected from patients living in SSP district. Three isolates were from patients living in Kwai Tsing district. Notably, the isolate obtained from the air sample collected in SSP district was also grouped in this clade. All isolates in this clade had a novel identical ST type (ST-1996 in PubMLST), shared a variable number of SNPs (31-2650 SNPs in 3856 cgMLST genes), and showed similar fragment profiles in the in silico PFGE result (grey block in Supplementary Figure 1). Among patients admitted in 2022, 90.9% had their isolates identified in this clade, indicating active transmission in the SSP and Kwai Tsing districts in 2022.
Figure 2. Phylogenetic analysis of 188 B. pseudomallei genomes (150 reference genomes and 38 genomes from isolates collected in this study). Maximum-likelihood phylogenies (mid-point rooted) of the SNPs of the 1646 single-copy cgMLST. Three monophyletic clades with more than one Hong Kong isolate identified are highlighted by colour blocks (grey: Clade 1, blue: Clade 2 and orange: Clade 3). Geographical location and sample host metadata of where the isolates were sampled were annotated and the sequence typing of the B. pseudomallei is denoted by circles next to the taxa. Bootstrap analysis of 1,000 replicates was performed and bootstrap values of selected nodes are shown (asterisk refers to 100%).
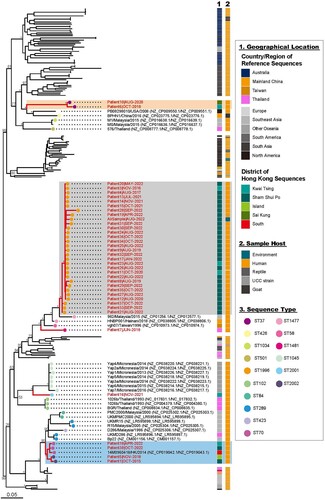
The second clade (coloured in blue in ) consisted of five B. pseudomallei isolates, four of which were from this study. Two were collected from patients living in the SSP district, one from the Kwai Tsing district, and one from the Island district. The remaining one was a previously published genome (BioProject ID: PRJNA342555), which was collected from a patient living in the Southern district, Hong Kong, in 2014. The isolates from this clade shared 96.28% to 98.15% SNP identities and showed similar fragment profiles in the in silico PFGE result (blue block in Supplementary Figure 1). They were separated into two ST types: ST-70, which has been identified in various locations in the Asia Pacific region, including Hong Kong, and a novel ST type, ST-2002.
The third clade (orange block in ) consisted of two B. pseudomallei isolates from patients living in Kwai Tsing (in 2018) and Sai Kung (in 2020) district. They were closest to the previously reported Isolate PB08298010 from the USA in 2008, with 91.78% to 92.54% genomic identities and an identical ST type (ST-37), which was the same as the animal isolates from a Hong Kong aquarium during 1985–1989 (whole genome sequences were not available) (Table S1). All isolates from this clade were included in the in silico PFGE analysis (orange block in Supplementary Figure 1), which showed that they were separated from the other two Hong Kong clusters described above, consistent with the phylogenetic analysis of the core genes.
Epidemiological and environmental investigation
As shown in , hospital admissions due to melioidosis were strongly associated with heavy daily rainfall and were almost exclusively aggregated during typhoon seasons. The statistical association between the disease and adverse weather events was determined using a Poisson regression model [Citation35,Citation37]. A significant correlation, in terms of odds ratio (OR), was found between melioidosis and the total weighted typhoon signal scores. Notably, the odds of case confirmation were the strongest in the third week after a typhoon [OR = 1.275 (95% CI = 1.076-1.451; p < 0.01)] (). Moreover, the correlation with prior rainfall within 7–28 days was also significant, although the strength of association was lower, with ORs ranging from 1.002-1.005 ().
Figure 3. Timeline showing the hospital admission date of melioidosis cases from 2015 to 2022 and the respective rainfall intensity and tropical cyclone warning signal (3 or higher) recorded during this period.
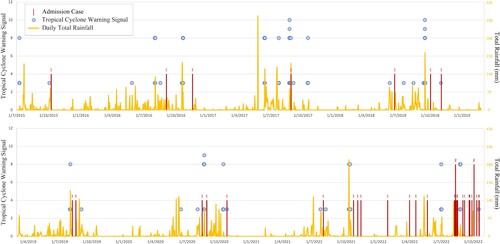
Table 2. Statistic correlation of prior typhoon and total rainfall with melioidosis cases in Kowloon West Region of Hong Kong (Jan 2015–Oct 2022) based on Poisson Regression analysis.
Out of the 38 patients, 30 (78.9%) lived in 25 separate apartment buildings within an estimated area of 2.50 km2 in the SSP district, KW Region (). To identify the transmission dynamics, 20 air samples and 72 soil samples were collected from a large building site, podium areas, and gardening areas between August 15 and October 5, 2022, near the residences of the recently infected patients.
Figure 4. The geographic location of the melioidosis cases in KW region from 2015 to 2022. The collection sites of the positive air sample and soil samples were also marked on the map. The wind data during the collection of the positive air sample on 15 August, 2022, 10:00–11:00 am was retrieved from Hong Kong Observatory.
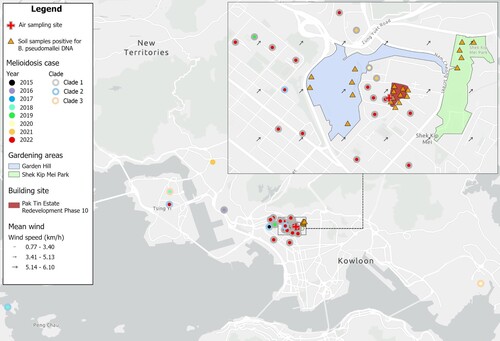
Notably, B. pseudomallei was successfully cultured from one (5.0%) of the air samples, which was collected on a second-floor podium 8.5 meters above the ground and 5 meters from a large building site (Pak Tin Re-development Phase 10) (red area in ). The collection date was August 15, 2022, five days after the attack of Typhoon Mulan, which was followed by the subsequent upsurge of melioidosis cases in the district.
Phylogenetic analysis showed that the air sample isolate, which was also typed as the novel ST-1996, was clustered in the same clade as the clinical outbreak strains in the SSP district ().
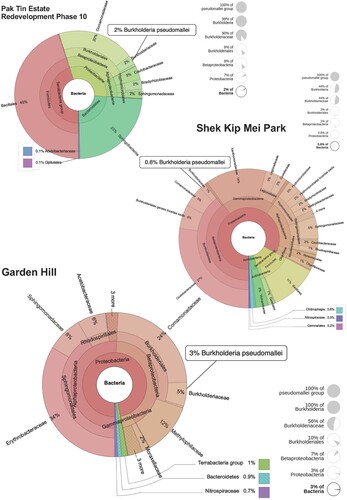
Although no positive culture of B. pseudomallei was obtained from soil, 16s rRNA gene sequencing detected B. pseudomallei DNA in 21/72 (29.2%) of the samples. Ten of these samples were collected from the building site, and 11 were from the gardening areas nearby, namely Garden Hill (n = 6) (blue area in ) and Shek Kip Mei Park (n = 5) (Green area in ). Quantitative analysis of 16s rRNA reads revealed that B. pseudomallei accounted for 3% of the bacterial population in the soil samples from Garden Hill, followed by 2% in the building site and 0.6% in Shek Kip Mei Park ().
Figure 5. Krona plots showing the abundance estimates of bacterial 16s rRNA sequences in soil samples collected from the building site (Pak Tin Estate Redevelopment Phase 10), Garden Hill and Shek Kip Mei Park. Interactive krona charts can be found in Figure S1–S3.
The gardening areas on August 15, 2022, from 10:00–11:00 am, had a mean wind direction from the southwest, and a mean speed of 5.14–6.10 km/hour (). It indicates that the wind came from the Garden Hill side during the collection of the positive air sample.
Area changes of vegetation region
The total area of the SSP district was calculated to be 9,516,363 m2. The vegetation region of the SSP district decreased from 1,725,033 m2 in 2016–1,562,778 m2 in 2022 (), representing a reduction of 162,255 m2. The vegetation in this area has been gradually declining over these years.
Discussion
There was a sudden upsurge of melioidosis cases observed from August to October 2022 in the KW Region, which serve 19% of the population in Hong Kong. Eighteen cases were identified within this two-month period, while the number of cases recorded in Hong Kong ranged from 4 to 17 every year in the past seven years (2015–2021) (). This clearly indicates an epidemic of the disease in Hong Kong. An outbreak of melioidosis was occurring in the SSP district of the KW region during the typhoon season in 2022.
Melioidosis is known as the “great mimicker” due to its non-specific clinical presentation. Patients with melioidosis present with features that are indistinguishable from community-acquired pneumonia (CAP), posing a significant diagnostic challenge to clinicians. Four patients presented with fever and respiratory symptoms and were treated empirically with amoxicillin-clavulanate as CAP. The diagnosis of melioidosis was not made until these patients experienced a recurrence of fever that required a second hospital admission 3–6 weeks later. Unfortunately, three of them eventually died of septicemic melioidosis.
In response to the outbreak, Ashdown’s agar has been added routinely for respiratory specimens of inpatients. Two out of 400 specimens were culture positive of B. pseudomallei (unpublished data). Clinicians should request specific B. pseudomallei culture for patients presenting with severe CAP or those with risk factors such as diabetes mellitus. Furthermore, laboratories in endemic areas should also consider routinely adding a selective medium of B. pseudomallei for sputum culture during typhoon season to improve diagnostic yield.
The increasing trend of melioidosis in Hong Kong is also observed in South Asian countries [Citation38]. A plausible explanation is climate change resulting in more frequent and intense weather events such as rainfall, tropical cyclones and flooding, all of which are associated with the occurrences and outbreaks of B. pseudomallei [Citation39–42]. A local study also revealed that the detection rate of B. pseudomallei from soil is positively correlated with ambient temperature and relative humidity [Citation14]. Prior to the unusual increase of melioidosis cases from August to October 2022, there were heavy rainfall and two typhoon attacks (Typhoon Mulan and Typhoon Ma-On) in early August 2022. Based on Poisson regression analysis, we found that melioidosis in Hong Kong was significantly correlated with prior typhoons and rainfall. The odds of hospital admission due to the disease were the strongest in the third week after a typhoon attack and increased by 27.5% when compared to non-exposure (). The time lag may reflect the chronological order of epidemiological causal inference given that the incubation period of melioidosis is usually 2–4 weeks. Patients who were exposed to contaminated aerosol during a typhoon are expected to develop symptoms and be admitted to the hospital 3–4 weeks later.
While melioidosis outbreaks after typhoon events have been reported in other endemic areas [Citation16,Citation42], none of these studies sampled viable ambient B. pseudomallei during typhoon season for phylogenetic analysis. In this study, we successfully cultivated B. pseudomallei from an air sample collected in close proximity to a cluster of melioidosis cases shortly after a typhoon attack. Core-genome phylogenetic analysis showed that the ambient isolate belonged to the same ST type and shared an identical phylogenetic placement with the outbreak cases, which provided direct evidence to support inhalation as the route of transmission during severe weather events. Inhalational melioidosis was also supported by circumstantial clinical evidence. Among the 18 patients who presented from August to October 2022, 13 (72%) had pneumonia, a higher incidence than other case series in endemic areas [Citation8,Citation43]. In addition, there were four melioidosis patients without any known risk factors, two of whom had severe necrotizing pneumonia requiring mechanical ventilation and intensive care. One patient, who was an air-conditioner technician, had a cough for one week before attending the hospital, and another patient had even more delayed presentation to the hospital after one month. Both patients eventually succumbed. Higher lethality of aerosol inhalation of B. pseudomallei was demonstrated by animal models [Citation10,Citation44]. Inhalation of aerosols containing a higher bacterial load during typhoons and rainstorms might be implicated in the increased severity of melioidosis, even in patients without predisposing conditions. The late presentation was likely due to the initial mild symptoms coupled with the lack of awareness of this potentially lethal infection, resulting in a worse outcome.
The sudden increase of melioidosis cases in such a small, urbanized area was highly unusual and prompted further epidemiological investigations. A large building site situated close to the residence of the SSP outbreak cluster (ST-1996) was identified as the place where the positive air sample for B. pseudomallei culture was obtained. Ten of 24 (41.7%) soil samples collected from the building site were positive for B. pseudomallei DNA. However, the construction work of the building site started in 2020, while the earliest case of ST-1996 in the SSP district was reported in 2016 (), indicating that the building site was not the only reservoir of this genotype. Further environmental investigation revealed more positive soil samples from the adjacent gardening areas, Garden Hill and Shek Kip Mei Park, signifying that B. pseudomallei is widely distributed in the SSP district. The soil sample with the highest abundance of B. pseudomallei DNA was collected from Garden Hill. Wind data retrieved from the Hong Kong Observatory showed that the wind came from the Garden Hill side during the collection of the positive air sample (i.e. August 15, 2022, 10:00–11:00 am) (). It was speculated that Garden Hill might be the source of the ambient B. pseudomallei isolate.
We further studied the changes in the vegetation region of the SSP district. The vegetation region area is the vertical projected area of vegetation (including leaves, stems and branches) on the ground [Citation45]. Studies have indicated that desertification increases the dust aerosol [Citation46–48], and unvegetated soil is more easily blown up by winds [Citation49,Citation50]. Analysis results from the satellite imagery showed that the vegetation region area of the SSP has been decreasing since 2015 due to continuous construction works for redevelopment. From 2016 to 2020, around 162,255.40 m2 of vegetated areas perished. The reduction in vegetation region, in conjunction with the heavy rainfall, accelerated the movement of B. pseudomallei from deeper soil layers to the surface for ease of aerosolization and further dispersal by strong winds during typhoons [Citation13,Citation39,Citation40].
The genetic diversity of B. pseudomallei populations was characterized based on MLST [Citation22,Citation51]. However, due to its high recombination rate, B. pseudomallei isolates might share the same MLST despite being genetically and geographically distinct [Citation52]. SNPs of the single-copy cgMLST genes identified in the whole-genome sequences have been used to study the phylogeny of B. pseudomallei, providing evidence of clustering of isolates with epidemiological links in high resolution [Citation53]. In this study, we used core-genome phylogenies of B. pseudomallei to study their emergence in the KW region of Hong Kong in the context of global diversity. Three local transmission clades of B. pseudomallei () in Hong Kong were revealed.
The first clade (ST-1996; ) demonstrated the genetic linkage between the air sample isolate and the recent clinical isolates in the SSP district. Notably, there was a relatively high prevalence of pneumonia observed in B. pseudomallei-infected patients in this clade, further supporting the potential airborne transmission route. Clinical isolates in this clade were sampled from 2016 to 2022 and, although they were closely related with the same genome structure, they demonstrated a significant number of SNPs (31-2,650 in 3,856 single-copy cgMLST computed using only the genomes within the clade), suggesting that a genetically variable population of B. pseudomallei may have been persistent in the nearby environment. B. pseudomallei is endemic in Hong Kong, and further genomic investigation could be helpful in elucidating the differences in virulence genes between the isolates in this clade and the others.
Two smaller clades of B. pseudomallei in Hong Kong might represent additional local transmissions of the bacterium. The second and third clades () included clinical isolates between 2014 and 2022 and those collected in 2018 and 2020, respectively. The three clades exhibited substantial differences in genomic structure and phylogenetic placement from each other, indicating different origins of their introductions to Hong Kong. Patients in these two clades were living in districts distantly separated for over 10 km from the KW district. High human mobility by the transportation system in Hong Kong and potential acquisition of infections in work or leisure places distant from home might explain the geographical variation in these clades. However, the long time separation between the cases made an investigation of the epidemiological links difficult.
Specifically, the two B. pseudomallei isolates in the smallest clade () had an identical ST type (ST-37) to the isolates collected from animals and environment sites of an aquarium and from patients diagnosed with melioidosis from 1975 to 1986 [Citation51]. This further supported that B. pseudomallei could be persistent in the environment for a long period of time. However, only the sequences of seven MLST genes were available for these isolates, and hence we were unable to elucidate the association between these isolates and the two isolates in the smallest clade at a higher resolution.
It is known that multiple genotypes of B. pseudomallei could exist in a single environmental site. For example, up to four genotypes of B. pseudomallei could be cultured from a single soil or bore water sample [Citation54,Citation55]. Therefore, a common environmental source of the infections cannot be completely ruled out. Enhanced surveillance of B. pseudomallei in soil, water and air in these districts will help to elucidate the areas of endemicity in Hong Kong and inform actions and policies to reduce bacterial colonization and human exposure to the contaminated sources.
One limitation of our study is its retrospective nature, which rendered the collection of epidemiological data incomplete for cases presented before August 2022. Secondly, a seroprevalence study was not conducted among citizens living in the SSP district, and the scale of the outbreak might be underestimated in patients with mild infections who did not seek hospital care. Nevertheless, the majority of patients presented with bacteremia, and the rise in melioidosis cases represented a significant outbreak. Another limitation is the inability to collect soil samples from deeper layers shortly after heavy rainfall. The highest concentration of B. pseudomallei was observed between 100 and 200 cm below the soil surface, as groundwater persistence is key to the occurrence of B. pseudomallei in soil [Citation56]. This may have lowered the yield of culture and metagenome study. Even so, based on the combined evidence obtained from this study, which included (i) the elevated odds ratios of the disease after typhoons and heavy rainfall, (ii) successful isolation of clonally related B. pseudomallei in an air sample collected near the residential areas of the patients, and (iii) the higher prevalence of pneumonia observed in the patients in this district, we postulate that the outbreak was caused by inhalation transmission of melioidosis after rainstorms and typhoons.
In conclusion, melioidosis is often a neglected infectious disease in Hong Kong until the recent upsurge of cases with significant morbidity and mortality. Here, we successfully isolated B. pseudomallei from air sampling, which was closely related to the clinical strains by cg-MLST. The detection of B. pseudomallei by 16S rRNA-gene analysis further supported the hypothesis of inhalational transmission of melioidosis after typhoons. Diagnosis of melioidosis remains exceptionally challenging due to the nonspecific clinical presentations. Clinicians should be aware of melioidosis, especially in patients with risk factors and after extreme weather events.
Supplemental Material
Download MS Excel (14.1 KB)Supplemental Material
Download PDF (238.9 KB)Acknowledgements
The authors wish to thank the hospital infection control team of Caritas Medical Centre for providing the air sampling machine, the microbiology laboratory staff of Princess Margaret Hospital and Public Health Laboratory Centre for isolating the clinical strains of B. pseudomallei and confirming the organism identity. The authors also wish to thank the Chief Infection Control Officer Office for providing the data on melioidosis in Hong Kong. Some computations in this research were performed using research computing facilities (high-performance computing clusters) offered by Information Technology Services, the University of Hong Kong.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Currie BJ, Dance DA, Cheng AC. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg. 2008 Dec;102(Suppl 1):S1–S4.
- Limmathurotsakul D, Golding N, Dance DA, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 2016 Jan 1;1(1):15008.
- Suputtamongkol Y, Chaowagul W, Chetchotisakd P, et al. Risk factors for melioidosis and bacteremic melioidosis. Clin Infect Dis. 1999 Aug;29(2):408–413.
- Hsueh PT, Huang WT, Hsueh HK, et al. Transmission modes of Melioidosis in Taiwan. Trop Med Infect Dis. 2018 Feb 28;3(1):26.
- Whitmore A. An account of a Glanders-like Disease occurring in Rangoon. J Hyg (Lond. 1913 Apr;13(1):1–34. 1.
- So SY, Chau PY, Leung YK, et al. First report of septicaemic melioidosis in Hong Kong. Trans R Soc Trop Med Hyg. 1984;78(4):456–459.
- Lui G, Tam A, Tso EYK, et al. Melioidosis in Hong Kong. Trop Med Infect Dis. 2018 Aug 25;3(3):91.
- Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis. 2010 Nov 30;4(11):e900.
- Currie BJ, Fisher DA, Howard DM, et al. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin Infect Dis. 2000 Oct;31(4):981–986.
- West TE, Myers ND, Liggitt HD, et al. Murine pulmonary infection and inflammation induced by inhalation of Burkholderia pseudomallei. Int J Exp Pathol. 2012 Dec;93(6):421–428.
- Currie BJ. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med. 2015 Feb;36(1):111–125.
- Ngauy V, Lemeshev Y, Sadkowski L, et al. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol. 2005 Feb;43(2):970–2.
- Currie BJ, Jacups SP. Intensity of rainfall and severity of melioidosis, Australia. Emerg Infect Dis. 2003 Dec;9(12):1538–1542.
- Lau SK, Chan SY, Curreem SO, et al. Burkholderia pseudomallei in soil samples from an oceanarium in Hong Kong detected using a sensitive PCR assay. Emerg Microbes Infect. 2014 Oct;3(10):e69.
- Currie BJ, Price EP, Mayo M, et al. Use of whole-genome sequencing to link Burkholderia pseudomallei from air sampling to mediastinal melioidosis, Australia. Emerg Infect Dis. 2015 Nov;21(11):2052–2054.
- Chen YL, Yen YC, Yang CY, et al. The concentrations of ambient Burkholderia pseudomallei during typhoon season in endemic area of melioidosis in Taiwan. PLoS Negl Trop Dis. 2014;8(5):e2877.
- Arslan F, Meynet E, Sunbul M, et al. The clinical features, diagnosis, treatment, and prognosis of neuroinvasive listeriosis: a multinational study. Eur J Clin Microbiol Infect Dis. 2015 Jun;34(6):1213–1221.
- Meumann EM, Novak RT, Gal D, et al. Clinical evaluation of a type III secretion system real-time PCR assay for diagnosing melioidosis. J Clin Microbiol. 2006 Aug;44(8):3028–3030.
- CLSI. Methods for antimicrobial dilution and disc susceptibility testing of infrequently isolated or fastidious bacteria. 3rd ed. M45. Wayne (PA): Clinical and Laboratory Standards Institute.
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410.
- Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009 Dec 15;10:421
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124.
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069.
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015 Jan 1;12(1):59–60.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014 May 1;30(9):1312–1313.
- Yu G, Lam TT, Zhu H, et al. Two methods for mapping and visualizing associated data on phylogeny using Ggtree. Mol Biol Evol. 2018 Dec 1;35(12):3041–3043.
- Heras J, Domínguez C, Mata E, et al. Gelj – a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics. 2015 Aug 26;16(1):270.
- Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995 Sep;33(9):2233–2239.
- Tobler WR. A computer movie simulating urban growth in the Detroit region. Econ Geogr. 1970;46(sup1):234–240.
- Paramasivam C, Venkatramanan S. An introduction to various spatial analysis techniques. In: Senapathi V, Prasanna MV, Sang YC, editor. GIS and geostatistical techniques for groundwater science. Amsterdam, Netherlands:Elsevier. 2019; p. 23–30.
- Zhao W, Zhong Y, Li Q, et al. Comparison and correction of IDW based wind speed interpolation methods in urbanized Shenzhen. Frontiers of Earth Science. 2022; 4(3A): 145–148.
- Deraman S, Wan Chik F, Muhammad M, et al. Case study: wind speed estimation of high-rise building using surface interpolation methods. Journal of Civil Engineering Research. 2014;4:145–148.
- Cellura M, Cirrincione G, Marvuglia A, et al. Wind speed spatial estimation for energy planning in Sicily: introduction and statistical analysis. Renewable Energy. 2008;33(6):1237–1250.
- Su HP, Chan TC, Chang CC. Typhoon-related leptospirosis and melioidosis, Taiwan, 2009. Emerg Infect Dis. 2011 Jul;17(7):1322–1324.
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, bacteria version 12.0; 2022.
- Liu X, Pang L, Sim SH, et al. Association of melioidosis incidence with rainfall and humidity, Singapore, 2003-2012. Emerg Infect Dis. 2015 Jan;21(1):159-162.
- Fang Y, Chen H, Li YL, et al. Melioidosis in Hainan, China: a restrospective study. Trans R Soc Trop Med Hyg. 2015 Oct;109(10):636–642.
- Currie BJ, Fisher DA, Howard DM, et al. The epidemiology of melioidosis in Australia and Papua New Guinea. Acta Trop. 2000 Feb 5;74(2-3):121–127.
- Suputtamongkol Y, Hall AJ, Dance DA, et al. The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int J Epidemiol. 1994 Oct;23(5):1082–1090.
- Birnie E, Biemond JJ, Wiersinga WJ. Drivers of melioidosis endemicity: epidemiological transition, zoonosis, and climate change. Curr Opin Infect Dis. 2022 Jun 1;35(3):196–204.
- Ko WC, Cheung BM, Tang HJ, et al. Melioidosis outbreak after typhoon, southern Taiwan. Emerg Infect Dis. 2007 Jun;13(6):896–898.
- Hantrakun V, Kongyu S, Klaytong P, et al. Clinical epidemiology of 7126 Melioidosis patients in Thailand and the implications for a National Notifiable Diseases Surveillance System. Open Forum Infect Dis. 2019, Dec;6(12):ofz498.
- Titball RW, Russell P, Cuccui J, et al. Burkholderia pseudomallei: animal models of infection. Trans R Soc Trop Med Hyg. 2008 Dec;102(Suppl 1):S111–S116.
- Deardorff JW. Efficient prediction of ground surface temperature and moisture, with inclusion of a layer of vegetation. Journal of Geophysical Research: Oceans. 1978;83(C4):1889–1903.
- Moridnejad A, Karimi N, Ariya PA. Newly desertified regions in Iraq and its surrounding areas: significant novel sources of global dust particles. J Arid Environ. 2015;116:1–10.
- Bergametti G, Marticorena B, Rajot J-L, et al. The respective roles of wind speed and green vegetation in controlling Sahelian dust emission during the wet season. Geophys Res Lett. 2020;47(22):e2020GL089761.
- Zhou C, Feng X, Huang Y, et al. The indirect impact of surface vegetation improvement on the climate response of sand-dust events in northern China. Atmosphere . 2021;12(3):339.
- Woodruff NP. How to control wind erosion. Department of Agriculture, Agricultural Research Service. Champaign, Illinois, USA. 1977; 354.
- Tsoar H, Møller JT. The role of vegetation in the formation of linear sand dunes. In: William GN, editor. Aeolian geomorphology. London, UK: Routledge; 2020. p. 75–96.
- Godoy D, Randle G, Simpson AJ, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, burkholderia pseudomallei and burkholderia mallei. J Clin Microbiol. 2003 May;41(5):2068–2079.
- Nandi T, Holden MT, Didelot X, et al. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res. 2015 Jan;25(1):129–141.
- Lichtenegger S, Trinh TT, Assig K, et al. Development and validation of a Burkholderia pseudomallei core genome multilocus sequence typing scheme To facilitate molecular surveillance. J Clin Microbiol. 2021 Jul 19;59(8):e0009321.
- Chantratita N, Wuthiekanun V, Limmathurotsakul D, et al. Genetic diversity and microevolution of Burkholderia pseudomallei in the environment. PLoS Negl Trop Dis. 2008;2(2):e182.
- Seng R, Saiprom N, Phunpang R, et al. Prevalence and genetic diversity of Burkholderia pseudomallei isolates in the environment near a patient’s residence in Northeast Thailand. PLoS Negl Trop Dis. 2019;13(4):e0007348.
- Pongmala K, Pierret A, Oliva P, et al. Distribution of burkholderia pseudomallei within a 300-cm deep soil profile: implications for environmental sampling. Sci Rep. 2022 May 23;12(1):8674.