Abstract
Monkeypox virus (MPXV) is a re-emerging zoonotic poxvirus responsible for producing skin lesions in humans. Endemic in sub-Saharan Africa, the 2022 outbreak with a clade IIb strain has resulted in ongoing sustained transmission of the virus worldwide. MPXV has a relatively wide host range, with infections reported in rodent and non-human primate species. However, the susceptibility of many domestic livestock species remains unknown. Here, we report on a susceptibility/transmission study in domestic pigs that were experimentally inoculated with a 2022 MPXV clade IIb isolate or served as sentinel contact control animals. Several principal-infected and sentinel contact control pigs developed minor lesions near the lips and nose starting at day 12 through 18 days post-challenge (DPC). No virus was isolated or viral DNA was detected from the lesions; however, MPXV antigen was detected by IHC in tissue from a pustule of a principal infected pig. Viral DNA and infectious virus were detected in nasal and oral swabs up to 14 DPC, with peak titers observed at 7 DPC. Viral DNA was also detected in nasal tissues or skin collected from two principal-infected animals at 7 DPC post-mortem. Furthermore, all principal-infected and sentinel control animals enrolled in the study seroconverted. In conclusion, we provide the first evidence that domestic pigs are susceptible to experimental MPXV infection and can transmit the virus to contact animals.
Disclaimer
As a service to authors and researchers we are providing this version of an accepted manuscript (AM). Copyediting, typesetting, and review of the resulting proofs will be undertaken on this manuscript before final publication of the Version of Record (VoR). During production and pre-press, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal relate to these versions also.Introduction:
Monkeypox virus (MPXV) is a re-emerging pathogen of the Orthopoxvirus (OPXV) genus in the Poxviridae family, which can spill over from a virus reservoir or intermediate host species into humans and cause disease. MPXV infection in humans produces illness reminiscent of smallpox infections, with clinical disease often presenting as fever and a characteristic maculopapular rash that progresses to vesiculopustular lesions [1, 2]. The severity of MPXV-produced disease is believed to be highly strain-dependent. Case fatality rates for clade I (Congo Basin) MPXV strains have been reported as high as 10% in unvaccinated people, while Clade II (West Africa) MPXV strains produce milder illness, with case fatality rates of less than 1% [3-5]. Unlike the closely related Variola minor and major viruses, the causative agents of smallpox, MPXV can infect a variety of animal species in addition to humans, and historically most human infections have been acquired through contact with either MPXV-infected non-human primates (NHPs) or rodents in endemic areas of Central and West Africa [2]. In 2003, sustained MPXV transmission was reported outside of the African continent for the first time, when the virus spread from imported African rodents (Gambian pouched rat, Cricetomys gambianus) to captive prairie dogs in the United States [18]. This resulted in 72 human infections, all of which occurred following direct contact with infected prairie dogs [6]. During the recent 2022 outbreak, human-to-human transmission was responsible for most of the approximately 90,000 cases recorded worldwide, although the initial transmission to humans is thought to have occurred through contact with an MPXV-infected animal. Therefore, identifying MPXV-susceptible animal species is paramount to predict, prevent and mitigate further zoonotic infections in humans.
Originally discovered in cynomolgus macaques (Macaca fascicularis) in 1958, MPXV is known to infect a variety of animal species, although the reservoir species for MPXV has yet to be identified [7]. Like humans, NHPs are considered incidental hosts for MPXV, as NHPs develop severe disease similar to that seen in humans [4]. MPXV has been isolated from chimpanzees and sooty mangabeys in the wild [8, 9], while macaques, marmosets, and baboons developed disease upon experimental inoculation [10-12]. Many rodent species are susceptible to MPXV infection, including giant pouched rats, Gambian pouched rats, rope squirrels, prairie dogs, woodchucks, and porcupines [13-19]. MPXV infection in prairie dogs resulted in: (i) necrotizing bronchopneumonia, conjunctivitis and tongue ulceration; (ii) positive virus isolation in lungs and tongue; and (iii) abundant viral antigen in surface epithelial cells of lesions in conjunctiva and tongue and in bronchial epithelial cells, macrophages, and fibroblasts in the lung [18]. Following experimental inoculation with either Clade I or II MPXV, prairie dogs develop signs of disease similar to those seen in humans, including the characteristic lesions [16-19]. By contrast, experimental inoculation of MPXV in most laboratory mouse strains produces mild or subclinical disease, although one recent report describes a mouse model that mimics the MPXV clade-dependent disease severity seen in humans [17,19, 22]. In addition to NHP and rodent species, MPXV or MPXV DNA has been detected in opossums, hedgehogs, and anteaters, raising concerns that other animal species may also be susceptible and could serve as virus reservoirs [15, 19].
The susceptibility of domestic animals to MPXV remains unknown. The domestic pig (Sus scrofa) is a common livestock amplification host for many zoonotic viruses, such as influenza A viruses, Japanese encephalitis virus, and Nipah virus. Pigs are also susceptible to at least one member of the Poxviridae family: swinepox virus. During initial isolation of MPXV, pig embryonic kidney (PEK) cells were used to amplify the virus, demonstrating in vitro susceptibility of pig cells to MPXV [20]. In addition, OPXV neutralizing antibodies were found in one pig from Central Africa during a large-scale serological survey [21]. One recent study reported that experimental Clade I MPXV inoculation of Siberian minipigs via the intranasal route produced no obvious clinical signs, although virus isolation and serology studies were not attempted [22].
In this study, we inoculated six 3-5 week-old piglets with Clade IIb MPXV via simultaneous intravenous, intradermal, and intranasal administration. Two days post-challenge (DPC), we introduced two sentinel piglets to determine whether MPXV can be transmitted to co-housed, in-contact pigs. Several pigs developed minor lesions near the lips and nose accompanied by MPXV antigen detection in skin samples collected upon necropsy. Infectious virus was isolated from the nasal and oropharyngeal swabs of two of the principally-challenged animals. Importantly, we were also able to isolate MPXV DNA in nasal and oropharyngeal swabs from all animals, including the sentinel contact control animals. The presence of anti-MPXV antibodies in principal-infected and sentinel animals confirmed pig-to-pig transmission. Therefore, this report provides the first evidence that pigs are susceptible to MPXV infection and can transmit the virus to contact animals.
Materials & Methods
Cells and viruses
Vero E6 cells (ATCC® CRL-1586™) were used for virus propagation and titration. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning, Corning NY, USA), supplemented with 2% fetal bovine serum (FBS, R&D Systems, Minneapolis, MN, USA) and penicillin/streptomycin (P/S, Gibco, Grand Island, NY, USA) and maintained at 37 °C in a 5% CO2 atmosphere. The hMPXV/USA/MA001/2022 (Lineage B.1, Clade IIb) isolate of MPXV was acquired from BEI Resources (Cat. #NR-58622) and used directly without further passaging for inoculation of animals. To determine the infectious titer of the original stock, 10-fold serial dilutions were performed on Vero E6 cells. Upon the appearance of cytopathic effect (CPE), the 50% tissue culture infectious dose (TCID50)/mL was calculated using the Spearman-Kaerber method [23].
Ethics statement
All animal studies and experiments were approved and conducted under the Kansas State University (KSU) Institutional Biosafety Committee (IBC, Protocol #1679) and the Institutional Animal Care and Use Committee (IACUC, Protocol #4824) in compliance with the Animal Welfare Act. All animal and laboratory work was performed in biosafety level-3 + and -3Ag (BSL-3+, BSL-3Ag) laboratories and facilities in the Biosecurity Research Institute (BRI) at KSU in Manhattan, KS, USA. Processing of inactivated formalin-fixed tissues occurred at BSL-2 as per Centers for Disease Control and Prevention (CDC) and BRI inactivation protocols.
Virus challenge of animals
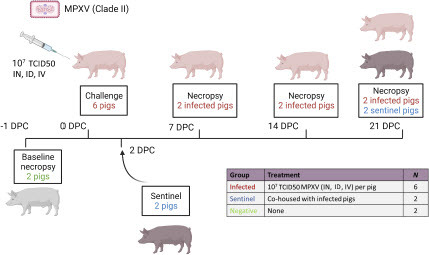
Ten male pigs, approximately 3-5 weeks old, were acquired from Oak Hill Genetics (Ewing, IL, USA) and allowed to acclimate for 3 days in the BSL-3Ag facility prior to the beginning of the study. One day before challenge, two pigs were euthanized to perform a full baseline post-mortem examination. Of the remaining eight pigs, six pigs were inoculated simultaneously via the intranasal, intradermal, and intravenous route with a 3 mL dose (1 mL per route) of MPXV at a concentration of 3 × 107 TCID50/mL. The remaining two pigs served as the contact sentinel controls and were separated from the principal-infected pigs until 2 DPC, when the animals were co-mingled (). Two principal-infected animals were each euthanized for post-mortem examination at 7, 14, and 21 DPC, while both contact sentinel controls were euthanized at 21 DPC.
Figure 1: Experimental design. Six pigs were inoculated with the MPXV hMPXV/USA/MA001/2022 (Lineage B.1, Clade IIb) isolate acquired from BEI Resources. A 3 ml dose of 1 × 107 TCID50 per animal was administered intranasal (IN), intradermal (ID), and intravenous (IV). At 2 days post-challenge (DPC), two contact sentinel control pigs were co-mingled with the six principally challenged animals. Daily clinical observations and body temperatures were recorded. Nasal, oropharyngeal, and rectal swabs as well as whole blood were collected at -1, 1, 3, 5, 7, 10, 14, 17, and 21 DPC. Oral fluids and serum were collected at -1, 7, 14, and 21 DPC. Post-mortem examinations were performed at 7, 14, and 21 DPC and results compared to baseline post-mortem examinations conducted on 2 additional negative control pigs at -1 DPC. BioRender.com was used to create the figure illustrations.
Clinical evaluations and sample collection
Pigs were observed daily for clinical signs, including depression, decreased appetite, respiratory signs, and lesions. Rectal temperatures were recorded daily. Nasal, oropharyngeal, and rectal swabs as well as whole blood were collected from animals at -1, 1, 3, 5, 7, 10, 14, 17, and 21 DPC. Swabs were placed in 1 mL of DMEM medium with P/S. Serum and oral fluids were collected before challenge and weekly following challenge at 7, 14, and 21 DPC. Blood was collected via venipuncture of the vena cava cranialis. Full post-mortem examinations were conducted at the indicated time points, and gross changes were noted. Haired skin was collected from the ears, eyelids, axillary region, inguinal region, and the ventral abdomen. Any grossly abnormal skin was also collected. Specimens from the respiratory tract included the nasal turbinates and ethmoturbinates, multiple sections of the trachea and bronchi and sections from all lung lobes. Lymphoid tissue collected included the nasopharynx, thymus, tonsil and numerous lymph nodes including the tracheobronchial, cranial mediastinal, retro-pharyngeal, mandibular, inguinal, superficial cervical, gastrohepatic, mesenteric and ileocecal lymph nodes. Visceral organs collected included the heart, liver, spleen, and kidney. Specimens from the gastrointestinal tract included the tongue, esophagus, stomach, and multiple locations throughout the small and large intestine. Miscellaneous tissues collected included the salivary glands, pancreas, adrenal glands, urinary bladder, skeletal muscle, bone marrow, olfactory bulb, brain and eye. Bronchoalveolar lavage fluid (BALF) of the left lung lobes was collected using 30-40 mL of DMEM. Cerebrospinal fluid (CSF), feces, and urine were also collected during post-mortem examination. All clinical samples were processed and stored at -80 °C until analysis. Tissue samples were formalin fixed or stored directly at -80 °C until analysis.
Histopathology
Tissues were fixed in formalin for 7 days, transferred to 70% ethanol (ThermoFisher Scientific, Liverpool, NY, USA), and stained with hematoxylin and eosin (H&E). Nasal cavity, rostral, middle and deep turbinates were decalcified with Immunocal™ Decalcifier (StatLab, McKinney, TX, USA) for 4 days at room temperature prior to H&E staining. Blinded examination was conducted by two veterinary pathologists.
Immunohistochemistry (IHC; Vaccinia-specific)
Immunohistochemistry (IHC) for detection of vaccinia virus B5R antigen (93% homologous to MPXV B5R homolog) was performed on the automated BOND RXm platform and using the Polymer Refine Red Detection kit (Leica Biosystems, Buffalo Grove, IL, USA). Following automated deparaffinization, four-micron formalin-fixed, paraffin-embedded tissue sections on positively charged Superfrost® Plus slides (VWR, Radnor, PA, USA) were subjected to automated heat-induced epitope retrieval (HIER) using a ready-to-use citrate-based retrieval solution (pH 6.0, Leica Biosystems) at 100 °C for 20 min. Subsequently, tissue sections were incubated with the primary antibody (rabbit polyclonal anti-vaccinia B5R (BEI Resources, Manassas, VA, NR-629) diluted 1:2,000 in Primary Antibody diluent (Leica Biosystems) for 30 min at ambient temperature followed by a polymer-labeled goat anti-rabbit IgG coupled with alkaline phosphatase (30 min). Fast Red was used as the chromogen (15 min), and counterstaining was performed with hematoxylin for 5 min. Slides were dried in a 60 °C oven for 30 min and mounted with a permanent mounting medium (Micromount®, Leica Biosystems). Sections derived from formalin-fixed, paraffin-embedded DF-1 cells infected with the modified vaccinia Ankara (MVA) virus were used as a positive assay control.
MPXV-specific RNAscope® in situ hybridization
For RNAscope® in situ hybridization (ISH), a commercially available and validated probe specific to MPXV (Advanced Cell Diagnostics (ACD), Newark, CA, USA, Cat. No. 534678) was used as previously described [24]. Sections of formalin-fixed paraffin-embedded tissues were generated as indicated above, and the RNAscope® ISH assay was performed using the RNAscope 2.5 LSx Reagent Kit (ACD) on the automated BOND RXm platform (Leica Biosystems). Following automated baking and deparaffinization, tissue sections were subjected to heat-induced epitope retrieval (HIER) using an EDTA-based solution (pH 9.0; Leica Biosystems) at 100 °C for 15 min, protease digestion using the RNAscope® 2.5 LSx Protease for 15 min at 40 °C, and incubation with a ready-to-use hydrogen peroxide solution for 10 min at room temperature. Slides were incubated with each probe mixture for 2 h at 40 °C, and the signal was amplified using a specific set of amplifiers (AMP1 through AMP6) as recommended by the manufacturer. The signal was detected using a Fast-Red solution for 10 min at room temperature. Slides were counterstained with a ready-to-use hematoxylin for 5 min, followed by five washes with 1X BOND Wash Solution (Leica Biosystems). Slides were finally rinsed in deionized water, dried in a 60 °C oven for 30 min, and mounted with Ecomount® (Biocare, Concord, CA, USA).
DNA extraction and quantitative PCR (qPCR)
The quantification of MPXV DNA was performed using a quantitative real-time PCR assay, i.e., G2R-G [25]. Briefly, frozen tissues were thawed and adjusted to 200 mg minced tissue/1 mL DMEM. Tissue homogenization was performed at 30 second intervals (4 pulses total for 2 minutes) at 30 Hz, using the Tissue Lyser LT (Qiagen, Hilden, Germany). Homogenates were pelleted at 3,000 g for 2 min.
DNA present in liquid (e.g. whole blood, swab samples, etc.) and homogenized samples was extracted using the GeneReach Total Nucleic Acid extraction kit on the Taco Mini Prime Purification System (GeneReach, Taichung City, Taiwan). Briefly, 100-200 µL of liquid sample was mixed with 500 µL lysis buffer containing 50 µL of magnetic beads, 150 μl of PBS (ThermoFisher Scientific), 40 μl proteinase K (Qiagen) and 200 μl of isopropanol (ThermoFisher Scientific) prior to the lysis and automated magnetic bead extraction.
For DNA extraction, tissue lysates were prepared using 250 μl of clarified tissue homogenate in 250 μl ATL buffer and 40 μl protease K. Tubes were incubated for 30 minutes at 56 °C on a heat block at 550 RPM shaking. ALT tissue lysates were incubated in the dark at room temperature for 3-5 days prior to processing DNA. One hundred μl of the tissue lysate was added to the lysis buffer and magnetic beads, followed by the addition of 100 μl of PBS and 200 μl of isopropanol.
DNA bound to beads was washed twice with the Wash buffer 1 (750 µL), once with the kit-supplied Wash buffer 2 (750 µL), once with absolute ethanol (750 µL) and dried at room temperature for 5 minutes. DNA was eluted in 100 µL of elution buffer.
The MPXV G2R-G assay was performed, using PerfeCTa® FastMix® II (Quantabio, Beverly, MA, USA) on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). A plasmid encoding the TNF receptor gene of MPXV was synthesized by GenScript (Piscataway, NJ, USA) and used as a positive control for all DNA extraction and qPCR reactions control. Serially diluted plasmids were used as reference samples for DNA copy numbers (see below). All qPCR reactions were undertaken with 5 µL of DNA template, 0.4 µM of primers and 0.2 µM of FAM probe (Integrated DNA Technology, Coralville, IA, USA) in a final reaction volume of 20 µL. Each reaction was performed in duplicate. Thermocycling parameters for the MPXV G2R-G assay were 95 °C for 5 min, followed by 45 cycles of 95 °C for 10 seconds and 60 °C for 1 minute. Negative and positive controls were included in each PCR run and consisted of molecular grade water and the MPXV-positive amplification control, respectively. A Ct cutoff of 38 was used for blood and swabs and 40 for tissues. These Ct cutoffs were selected based on the analytical limits of the qPCR assay and recommendations from the CDC website for this assay [26]. Samples with both PCR replicates at or below the Ct cutoffs were interpreted as positive. Samples with one of both PCR replicates at or below the Ct cutoffs were interpreted as suspect.
The screening of MPXV DNA in skin lesion swabs and samples was undertaken with a qPCR assay targeting the E9L gene [27] according to CDC’s recommendations [28].
Virus isolation from clinical samples
Nasal and oral swab samples were briefly vortexed before adding 500 μl to 6-well plates of Vero E6 cells. Cells were incubated at 37 °C for 2 hours before the removal of inoculum and washing cells once with phosphate buffered saline (PBS). Then, 2 mL of 2% FBS DMEM media with 1% P/S was added per well and plates were incubated for 4 days at 37 °C.
To visualize MPXV-specific CPE, plates of cells were fixed with 80% acetone for 10 minutes and washed twice with PBS. Next, 1 mL of rabbit anti-vaccinia virus (VACV) polyclonal antibody (PA1-7258, Invitrogen, Waltham, MA, USA) diluted 1:250 in 1% bovine serum albumin (BSA) in PBS was added per well, and plates were incubated for 1 hour at room temperature (RT). Plates were then washed 3 times with PBS-T (0.1% Tween-20 in PBS) before adding 1 mL of Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) diluted 1:1000 in 1% BSA. After 1 hour of incubation at RT, plates were washed another 3 times and plaques visualized using an EVOS fluorescent microscope.
Indirect enzyme-linked immunosorbent assay (ELISA)
To detect MPXV binding antibodies in sera, indirect ELISAs were performed using purified recombinant A35 and H3 MPXV antigens (Sino Biological Inc., Beijing, China). Briefly, 96-well Maxisorp immunoassay plates (ThermoFisher) were coated with 300 ng of either A35 or H3 antigen per well in PBS and incubated at 4 °C overnight. After blocking with 5% skim milk in PBS-T, plates were incubated for 1 hour with sera diluted in 1% skim milk in PBS-T. Plates were then washed 3 times with PBS-T before adding a 1:5000 dilution of goat anti-pig IgG conjugated with HRP (Abcam, Cambridge, UK). After 1 hour, plates were again washed 3 times with PBS-T and 100 μl of TMB substrate (Abcam) were added per well. Plates were incubated for 5 minutes before adding 100 μl of stop solution (Abcam). The OD of the ELISA plates were read at 450 nm on an ELx808 BioTek plate reader, and a sample was considered positive when the OD value was higher than the mean OD of naïve serum plus 3 standard deviations.
Whole-virus ELISAs were also performed using sample dilutions of 1:100, 1:200, and 1:400 as previously described [29]. Briefly, 0.01 µg/well of crude vaccinia virus (Western Reserve strain) diluted in carbonate buffer was used to coat half of a microtiter plate, while the other half of the plate was coated with BSC-40 cell lysate. A cut-off value (COV) was generated by calculating the average OD value from the virus half of the plate and subtracting the average plus two standard deviations of the corresponding sample in the cell lysate half of the plate. All samples were run in duplicate. An animal was considered positive if the sample OD value was above the COV.
Detection of MPXV binding antibodies by immunofluorescence
The 96-well plates of Vero E6 cells were infected with MPXV at a multiplicity of infection (MOI) of 1 and incubated for 48 hours at 37 °C before being fixed with 80% acetone. Wells were rehydrated with PBS, and 50 μl of pig serum samples (diluted 1:10 in 1% BSA) were added to each well. As a positive control, 50 μl of rabbit anti-VACV polyclonal antibody (PA1-7258, Invitrogen) diluted 1:250 in 1% BSA were added to wells. After incubation for 1 hour at RT, plates were washed 3 times in PBS-T. Plates were then incubated with 1 mL of a 1:250 dilution of goat anti-porcine IgG-AF488 (Southern Biotech, Birmingham, AL, USA) or biotinylated Protein A conjugated with streptavidin, DyLight 488 (Vector Laboratories, Newark, CA, USA). Positive control wells were incubated with either Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) or Protein A. After washing 3 times with PBS-T, Vectashield Plus antifade medium with DAPI (Vector Laboratories) was added to counterstain and plates were visualized using an EVOS fluorescent microscope.
Results
Pigs remain subclinical following challenge with MPXV
Clinical signs that included activity level, appetite, respiratory signs, and skin lesions were monitored daily, and rectal temperatures were also recorded daily. Body temperatures for all animals remained within the normal range throughout the course of the study (Suppl. Fig. 1). Beginning at 12 days post-challenge (DPC), some minor skin lesions were observed in both the principally-infected and sentinel contact control animals. Lesions were observed near the lips and nose of four pigs from 12 DPC through 18 DPC (Suppl. Fig. 1). These lesions presented as red macules on three animals, and a singular large vesicle on the nose of a fourth animal (Suppl. Fig. 1). Observed lesions were swabbed for subsequent qPCR; however, no viral DNA could be detected. No other clinical signs were observed throughout the remainder of the study.
Comprehensive post-mortem exams were performed on all pigs. No significant gross lesions were seen in visceral organs; and lesions were restricted to the skin. Regions of hyperemia, erosions, ulcers, and scabs were collected from each animal when present. At 7 DPC, epidermal changes were only observed in one of the two principal-infected pigs (ID #23). The upper right eyelid had a 0.5 cm linear crusty lesion near the medial canthus. This pig also developed two red, non-raised circular foci approximately 1 cm in diameter on the left lateral side of the nose. These regions consisted of hyperemia and mild thickening of the epidermis and hyperkeratosis, with rare intracorneal pustules at the level of the eyelid.
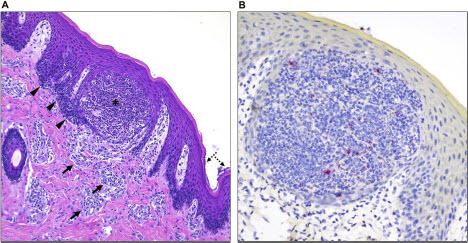
At 14 DPC, skin lesions were observed in both pigs examined. One pig (#17) had a 0.5 cm pustular to ulcerated focus on the ventral chin (Suppl. Fig. 3A), delimited by a dark-red congested rim (Suppl. Fig. 3A insert). Overall, the microscopic alterations included mild multifocal pustular, hyperplastic and lymphohistiocytic perivascular dermatitis with occasional epidermal ulceration (Suppl. Fig. 3B). Mononuclear inflammatory cells infiltrated the superficial dermis underlying the ulcerated epidermis and also delimiting dermal blood vessels. The adjacent epidermis was hyperplastic (acanthosis) and the stratum corneum was occasionally covered with small serocellular crusts containing coccoid and rod-shaped bacteria. At 14 DPC, pig #20 developed wet eyelids and slight hyperemia within the left conjunctiva and the surrounding skin. Small 2 mm crusts occurred on the lower and upper eyelids above and below the lashes. Furthermore, formation of epidermal pustules was noted (). Small amounts of intracytoplasmic viral antigen in degenerate inflammatory cells within a single pustule in pig #20 were identified by immunohistochemistry using a polyclonal anti-vaccinia B5R antibody (). However, no viral mRNA transcripts were detected via in situ hybridization using a MPXV-specific probe.
Figure 6. Histological alterations in the skin and immunohistochemical detection of MPXV antigen in one of the infected pigs. (A) Histologic alterations are characterized by epidermal pustule formation (*), hyperplasia of the adjacent epidermis (arrowheads) and superficial perivascular lymphohistiocytic and eosinophilic dermatitis (arrows). The corneal layer of the epidermis was occasionally colonized by coccoid bacteria (dashed arrows). (B) Within this single pustule, sporadic degenerate inflammatory cells contain minimal intracytoplasmic viral antigen (red). No viral antigen was detected within keratinocytes or other cell types. H&E, 200X total magnification; Fast Red, 400X total magnification.
Among the animals euthanized at 21 DPC (n = 4, corresponding to two principal infected [#21 and #24] and two sentinel pigs [#16 and #18]), three pigs developed skin alterations. Consistent with the skin lesion on the left lateral chin (Suppl. Fig. 1C) observed at 12 DPC, pig #24 developed pustular dermatitis with epidermal acanthosis (Suppl. Fig. 3C). Lesions from the ventral abdomen had similar erosive dermatitis. Interestingly, pig #18 developed a 1.5 cm diameter vesicular lesion in the dorsal nasal planum (11 DPC). This lesion rapidly progressed into a ruptured vesicle in the superficial epidermis (12 DPC) that developed into an erosion with transient crust formation (Suppl. Fig. 1D). The lesion rapidly resolved and was not apparent at the time of necropsy (21 DPC). Skin lesions appreciated grossly in pig #16 at 21 DPC included multifocal small crusts on the upper eyelid, and histologically were characterized as multifocal suppurative folliculitis and dermatitis with mild-to-moderate epidermal acanthosis. No viral antigen or DNA in situ was identified in this group.
MPXV virus and/or viral DNA can be isolated from the upper respiratory tract of all pigs
Nasal, oral, and rectal swabs were collected regularly throughout the course of the study in order to determine MPXV virus shedding in pigs. MPXV DNA was detected in nasal swabs from all six principally-infected pigs at 1 DPC, and MPXV DNA continued to be detectable up to 7 DPC in four animals (A). MPXV DNA levels increased over time in two of the four animals with persistent nasal shedding, with a peak titer of 2.20 × 107 DNA copies/swab detected at 7 DPC in one animal (pig #24). In addition, MPXV DNA was detected in the nasal swabs of both sentinel contact animals at 7 and 10 DPC. Four out of six principally-infected animals and both sentinel contact control animals also had detectable MPXV DNA in oral swabs from 3 DPC to 14 DPC, although DNA levels were generally lower than those observed in nasal swabs (B). Low levels of MPXV DNA could be detected in the rectal swabs of three principally-infected animals at 1 DPC, though no other rectal samples collected from the principal or sentinel animals tested positive throughout the remainder of the study (data not shown).
Figure 2: Viral DNA shedding of MPXV-infected pigs. qPCR was performed on nasal (A, C) and oral (B, D) swabs collected from principal-infected and sentinel contact control pigs at indicated the timepoints. Mean (n = 2) viral DNA copy number per swab is reported based on the detection of the MPXV TNF receptor gene (crmB). The dotted line indicates the limit of detection for the assay. (A, B) Numbers indicate the ID code of the individual pig. An open symbol indicates samples with only one of two qPCR reactions positive. (C, D) Line graphs display the mean and error bars indicate the standard deviation.
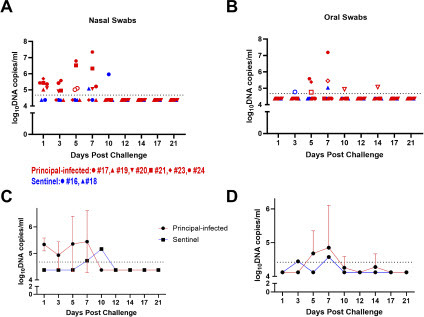
Virus isolation was attempted for any sample with an MPXV DNA copy number above 2 × 105 genomic copies/swab. To distinguish MPXV-specific CPE from toxicity caused by other contaminants in the clinical samples, we fixed cells with 80% acetone and stained with a rabbit polyclonal antibody against VACV (PA1-7258) that is known to be cross-reactive with MPXV [28]. We identified MPXV-specific staining in four clinical samples: three nasal samples from 1 and 5 DPC and one oral sample from 7 DPC from two different principal infected pigs (Suppl. Fig. 2).
In order to determine whether MPXV could spread beyond the upper respiratory tract of animals, blood and oral fluids were periodically collected throughout the study and a full panel of tissues was collected at post-mortem examinations. No MPXV DNA could be detected in the blood or oral fluids of any animal at any point during the study. Only two animals had detectable MPXV DNA in the nasal ethmoturbinates at 7 DPC, and one additional animal had MPXV DNA in the skin at 7 DPC (Suppl. Table 1).
Pigs develop an antibody response that is reactive to MPXV
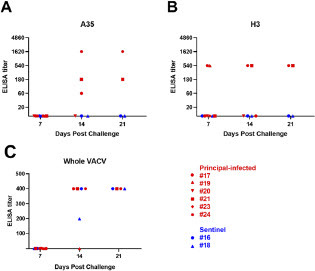
Serum samples were collected from each animal weekly for serological testing. Indirect ELISA tests were used to detect anti-MPXV IgG against recombinant A35 and H3 MPXV antigens. A35 (homologous to VACV A33) and H3 are envelope proteins found on the surface of mature virions and are known to be targets for neutralizing antibodies in VACV-vaccinated individuals [30-32]. Two of the six principal-infected animals developed IgG antibodies binding either A35 or H3 by 7 DPC, and two additional animals had developed detectable binding antibodies at 14 DPC (A and 3B). While binding antibodies could not be detected in the remaining two principally-infected animals, both of these animals were euthanized prior to the end of the study. Neither sentinel animal had developed detectable binding antibodies to either H3 or A35 antigens by the end of the study. In addition to performing MPXV single-antigen ELISA assays, we also performed whole-virus ELISA assays using VACV. Using this assay, anti-OPXV antibodies were detected in three principal-infected animals and both sentinel contact animals, starting at 14 DPC (C).
Figure 3: Detection of binding antibodies targeting MPXV. Indirect ELISA using purified recombinant MPXV A35 (A) and H3 (B) antigens were used to detect MPXV-specific binding antibodies in pig sera. Serum was diluted in 3x serial dilutions. An ELISA titer was considered positive when the OD450 was greater than 3 standard deviations above the mean of naïve animal sera run concurrently. (C) Indirect ELISA using crude whole VACV. Plates were coated with either VACV or BSC-40 cell lysate. Sera was diluted 1:100, 1:200, and 1:400. An ELISA titer was considered positive when the average OD450 among two replicates was greater than 2 standard deviations above the mean of corresponding samples in the cell lysate plate.
We next examined the total IgG response of the pig sera against MPXV-infected Vero E6 cells. With over 200 viral genes, MPXV virions contain dozens of possible antigens. In addition, two infectious forms of the virus, the intracellular mature virion and the extracellular enveloped virion, are produced during the infectious cycle that each display a separate set of surface proteins [33]. To capture the full IgG antibody response to MPXV, we infected Vero E6 cells with MPXV and incubated with serum from MPXV-infected pigs, followed by secondary staining with a goat anti-pig IgG antibody. Low levels of antibodies could be detected in both principally-infected animals that had previously tested negative by ELISA (). In addition, antibodies were also detectable in one of the two sentinel animals ().
Figure 4: Immunofluorescent detection of MPXV-specific total IgG. Representative images of stained cells. Naïve Vero E6 cells were infected with MPXV at an MOI of 1 and incubated for 48 hours before fixation with 80% acetone. Pig sera was diluted in PBS with 1% BSA and added to fixed wells. Goat anti-pig IgG secondary antibody conjugated to Alexa-Fluor 488 was used to stain wells, and wells were visualized at 10x magnification using an EVOS fluorescence microscope. Naïve serum collected at -1 DPC was used as a negative control, and a rabbit anti-vaccinia polyclonal antibody was used as a positive control. Scale bars represent 400 µm.
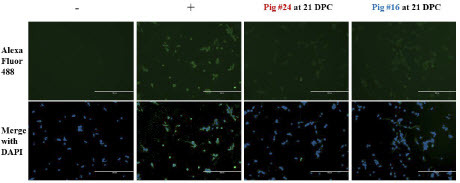
Table 1: Summary of indirect immunofluorescence staining of infected Vero cells to detect MPXV-reactive IgG in pig serum. Detection of IgG was based on goat anti-pig IgG polyclonal antibodies.
We further examined the total antibody response to MPXV using IF where antibodies bound to MPXV antigens were detected by Protein A. Protein A binds strongly to pig total IgG. In addition, Protein A has been reported to weakly bind human IgM and IgA as well, though binding affinity for porcine IgM and IgA has not been tested. We incubated MPXV-infected cells with serum from MPXV-infected pigs as indicated above, followed by Protein A. Using this method, antibodies could be detected in four out of six principal-infected animals and both sentinel animals by 14 DPC (; ).
Figure 5: Detection of MPXV-specific antibodies using Protein A. Vero E6 cells were infected with MPXV at an MOI of 1 and incubated for 48 hours before fixation with 80% acetone. Pig sera was diluted in PBS with 1% BSA and added to fixed wells. Biotinylated Protein A conjugated to streptavidin-DyLight488 was used to stain wells, and wells were visualized at 10x magnification using an EVOS fluorescence microscope. Scale bars represent 400 µm. (A) Representative images of stained cells. Naïve serum collected at -1 DPC was used as a negative control, and a rabbit anti-vaccinia polyclonal antibody was used as a positive control. (B) Images from sentinel pig #18, collected at each indicated timepoint.
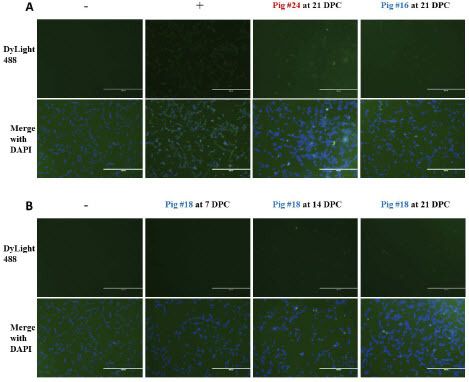
Table 2: Summary of indirect immunofluorescence staining of infected Vero cells to detect MPXV-reactive antibodies in pig serum. Detection of antibodies was based on Protein A.
Discussion
In this study, we investigated the susceptibility of the domestic pig (S. scrofa) to Clade II MPXV infection using an isolate from a human case during the recent 2022 outbreak. Until this outbreak, zoonotic spread was thought to be responsible for the majority of reported MPXV infections in humans. MPXV is known to infect a wide variety of NHP and rodent species in endemic areas of Central and West Africa, but the susceptibility of many common domestic species to MPXV remains unknown. In addition, while many previous studies have evaluated possible laboratory animal models for MPXV infection, many of these studies exclusively used Clade I MPXV strains. One early study using Clade I MPXV reported that rubbing virus into the skin of pigs did not result in disease [20, 34]. A more recent report evaluated MPXV infection in miniature pigs, which concluded that pigs remained subclinical following intranasal Clade I MPXV challenge infection [22]. In our study, to assess overall susceptibility and possible utility as an animal model, we inoculated pigs with a high dose of MPXV via three different inoculation routes simultaneously: intranasal, intradermal, and intravenous. While our findings are in concordance with the previous reports that MPXV infection of pigs is subclinical, we observed evidence of productive infection of the upper respiratory tracts of pigs, seroconversion, and transmission to contact sentinel control animals.
This study provides clear evidence that MPXV can productively replicate in the upper respiratory tract of pigs. Several principal-infected and sentinel contact controls pigs developed minor lesions near the lips and nose starting at day 12 through 18 DPC. None of the lesions tested positive for MPXV DNA via qPCR, so these lesions may be incidental, though tissue samples collected from one pustule in pig #20 reacted with an anti-vaccinia antibody following IHC staining. However, MPXV DNA could be isolated until 7 DPC in nasal swabs collected from four out of six challenged animals. While DNA detected at early timepoints post-challenge could be attributed to remnants of virus inoculum, DNA levels increased over time by approximately 100-fold in two animals and remained stable through 7 DPC in the remaining two animals. Oral swabs collected from one of these animals also demonstrated a nearly 100-fold increase in MPXV DNA from 5-7 DPC. MPXV stability in bodily fluids has not been fully characterized, though one recent study reported that MPXV infectious virus could no longer be detected after 6 days of incubation in sheep blood at 37 °C [35]. We detected infectious virus in nasal and oral swabs collected as late as 5-7 DPC. Although our sample size was small, we conclude from our results that MPXV infection in pigs can be productive.
The tissue tropism of MPXV in pigs remains to be fully characterized, although upper respiratory tissues are likely the major source of MPXV replication. MPXV DNA was never detected in blood samples despite the initial intravenous challenge, and rectal swabs were also negative beyond 1 DPC. The majority of organs tested upon necropsy did not contain detectable MPXV DNA. However, at 7 DPC, MPXV DNA was detected in the nasal ethmoturbinates or skin of two principal-infected animals. MPXV DNA load in nasal and oral swabs peaked at 7 DPC and became undetectable in samples collected beyond 14 DPC. This is consistent with previous MPXV challenge studies in prairie dogs and macaques, in which viral titers peaked in oral samples collected from 6-13 DPC [36-41]. Therefore, it is possible that the remaining pigs had cleared the infection by the time necropsies were conducted at 14 and 21 DPC.
Inoculation of pigs with MPXV was able to induce a humoral response in all pigs in this study. The two principal-infected animals with the highest MPXV DNA titers both had the highest antibody titers as detected by ELISA, suggesting that a proportionally greater humoral response was mounted in response to increased viral replication in these animals. Previous MPXV challenge studies in macaques and baboons reported the appearance of anti-MPXV antibodies at around 8 DPC [12, 42], while in prairie dogs, antibodies first appeared as late as 12 DPC [39]. Similarly, we could not detect antibodies in some of our pigs until 14 DPC. One major limitation of this study is that, while the IgG response was characterized by both ELISA and IF, IgM and IgA titers were not directly measured. As our results indicate that MPXV infection in pigs is largely confined to the upper respiratory tract, the IgA response might be a particularly important factor of the immune response to MPXV in pigs.
One important finding from our study is that MPXV is able to spread to other pigs. Traditional ring vaccination strategies for both smallpox and mpox outbreaks in humans have relied on the assumption that these viruses cannot be transmitted in the absence of clinical signs of disease [43.]. Nevertheless, during the 2022 mpox outbreak, some transmission was reported to occur before the onset of symptoms [44]. Among the pigs in our study, MPXV DNA could be detected in both nasal and oral swabs collected from both sentinel contact control animals. In addition, both sentinel animals developed anti-MPXV antibodies by 14 DPC. While MPXV binding antibodies were not detectable via single-antigen ELISA for either sentinel animal, both sentinel animals were positive using a whole-virus ELISA as well as by IFA. In one sentinel contact animal (pig #16), we detected more than 1.9 × 105 MPXV DNA copies in the nasal swab collected at 10 DPC. Antibodies began to appear in this animal as early as 7 DPC. In the other sentinel animal (pig #18), relatively lower levels of MPXV DNA were detected in both the nasal and oral swabs collected at 7 DPC, and we could only detect anti-MPXV antibodies in this animal starting at 14 DPC. No MPXV DNA could be detected in either animal after 10 DPC, which may indicate that infection was transient. Nevertheless, the possibility that MPXV infection can spread among pigs raises concerns that the virus can be maintained in livestock populations, which may increase the likelihood of further zoonotic infections in humans. Interestingly, minimal viral antigen was identified within inflammatory cells in a single pustule from one of the principal-infected pigs despite the development of mild pustular and hyperplastic dermatitis in several of the infected pigs. While the period of time elapsed between the initial formation of these skin lesions and postmortem evaluation could explain the low detection rate, this finding suggests that the skin alterations seem to be directly associated with MPXV infection.
While our study does demonstrate the susceptibility of the domestic pig to MPXV, the natural route of MPXV infection from pig to pig remains unclear. We experimentally infected pigs using three different routes of inoculation and a high dose of MPXV. Our results suggest that the principal challenged animals were able to transmit MPXV to the sentinel animals, as evidenced by viral DNA found in the clinical swabs and seroconversion in both sentinel animals. However, it is unclear how the virus was transmitted to these pigs. Furthermore, since only low levels of viral DNA and no infectious virus could be detected from either sentinel animal, it is less likely that sentinel animals could further transmit the virus to other naïve animals. Further studies are needed to determine the exact transmission route of MPXV between co-housed pigs.
In conclusion, our study demonstrates for the first time that pigs are susceptible to productive infection with Clade II MPXV and can spread infection to other pigs. The 2022 strain used in this study has been reported to cause milder disease and reduced infectious titers than Clade I and other Clade II strains of MPXV in mouse models [17, 19, 45]. Therefore, future studies should evaluate the pathogenesis of representatives of Clade I and more virulent Clade II strains of MPXV in pigs. In addition, widespread serological surveys are needed to address the possibility of natural MPXV infection in domestic swine populations. Most surveys to date have focused on rodent and NHP species [29, 46-47], though one survey did collect a single sample from a pig that subsequently tested positive for anti-OPXV antibodies [20]. Future large-scale serological surveys in pigs and other livestock species should be conducted to better understand the risk of MPXV spread in agricultural settings.
Disclosure:
The J.A.R. laboratory received support from Tonix Pharmaceuticals, Genus plc, Xing Technologies, and Zoetis, outside of the reported work. J.A.R. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by Kansas State University. The other authors declare no competing interests.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The conclusions in this report are those of the authors and do not necessarily represent the views of the USDA. USDA is an equal opportunity provider and employer. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official views, position, or policy of the Centers for Disease Control and Prevention or the United States federal government, and no official endorsement should be inferred.
Author Contributions
JAR, LA, IM, NO, WW, JDT, and EM were involved in the study conception and design. EM, CL, JDT, NNG, IM, FMF, CM, DB, TK, KC, VB, DM, BA, JS, JBD, WW, NO, and JAR were involved in the data collection. EM, JDT, FMF, NNG, NO, and JAR were involved in data analysis, and interpretation of results. EM prepared the first draft of the manuscript. All authors revised and reviewed the manuscript and agree to be accountable for the content of the work.
Acknowledgements:
We thank the staff of KSU Biosecurity Research Institute and KSU Veterinary Diagnostic Histopathology Laboratory as well as the Louisiana Animal Disease Diagnostic Laboratory Histology and Immunohistochemistry section. We are thankful to Yonghai Li, Eulim Lyoo, Patricia Assato, Isaac Fitz and Bailey Roberts from KSU for technical support. We also thank Bryon Nelson and Katie Knapek from the National Bio- and Agro-defense Facility, Agricultural Research Service, United States Department of Agriculture, for technical support. The following reagent was obtained through BEI Resources, NIAID, NIH: Polyclonal Anti-Vaccinia Virus (WR) B5R Protein, (antiserum, Rabbit), NR-629. Funding for this study was provided through grants from the National Bio and Agro-Defense Facility (NBAF) Transition Fund from the State of Kansas, the USDA Animal Plant Health Inspection Service’s NBAF Scientist Training Program, the AMP and MCB Core of the Center on Emerging and Zoonotic Infectious Diseases (CEZID) of the National Institutes of General Medical Sciences under award number P20GM130448, and the USDA NACA #58-3022-3-004.
References
- Foster, S.O., et al., Human monkeypox. Bull World Health Organ, 1972. 46(5): p. 569-76.
- Damon, I.K., Status of human monkeypox: clinical disease, epidemiology and research. Vaccine, 2011. 29 Suppl 4: p. D54-9.
- Likos, A.M., et al., A tale of two clades: monkeypox viruses. J Gen Virol, 2005. 86(Pt 10): p. 2661-2672.
- Cann, J.A., et al., Comparative pathology of smallpox and monkeypox in man and macaques. J Comp Pathol, 2013. 148(1): p. 6-21.
- Chen, N., et al., Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology, 2005. 340(1): p. 46-63.
- Reed, K.D., et al., The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med, 2004. 350(4): p. 342-50.
- P., v.M., et al., A pox-like disease in cynomolgus monkeys. 1959, Acta Pathol. Microbiol. Scand. p. 156–176.
- Radonić, A., et al., Fatal monkeypox in wild-living sooty mangabey, Côte d'Ivoire, 2012. Emerg Infect Dis, 2014. 20(6): p. 1009-11.
- Patrono, L.V., et al., Monkeypox virus emergence in wild chimpanzees reveals distinct clinical outcomes and viral diversity. Nat Microbiol, 2020. 5(7): p. 955-965.
- Mucker, E.M., et al., Effect of Monkeypox Virus Preparation on the Lethality of the Intravenous Cynomolgus Macaque Model. Viruses, 2022. 14(8).
- Mucker, E.M., et al., Susceptibility of Marmosets (Callithrix jacchus) to Monkeypox Virus: A Low Dose Prospective Model for Monkeypox and Smallpox Disease. PLoS One, 2015. 10(7): p. e0131742.
- Heberling, R.L. and S.S. Kalter, Induction, course, and transmissibility of monkeypox in the baboon (Papio cynocephalus). J Infect Dis, 1971. 124(1): p. 33-8.
- Silva, N.I.O., et al., Here, There, and Everywhere: The Wide Host Range and Geographic Distribution of Zoonotic Orthopoxviruses. Viruses, 2020. 13(1).
- Khodakevich, L., Z. Jezek, and K. Kinzanzka, Isolation of monkeypox virus from wild squirrel infected in nature. Lancet, 1986. 1(8472): p. 98-9.
- Hutson, C.L., et al., Monkeypox zoonotic associations: insights from laboratory evaluation of animals associated with the multi-state US outbreak. Am J Trop Med Hyg, 2007. 76(4): p. 757-68.
- Falendysz EA, Lopera JG, Lorenzsonn F, Salzer JS, Hutson CL, Doty J, Gallardo-Romero N, Carroll DS, Osorio JE, Rocke TE. Further Assessment of Monkeypox Virus Infection in Gambian Pouched Rats (Cricetomys gambianus) Using In Vivo Bioluminescent Imaging. PLoS Negl Trop Dis. 2015 Oct 30;9(10):e0004130. doi: 10.1371/journal.pntd.0004130. PMID: 26517839; PMCID: PMC4627722.
- Americo, J.L., P.L. Earl, and B. Moss, Virulence differences of mpox (monkeypox) virus clades I, IIa, and IIb.1 in a small animal model. Proc Natl Acad Sci U S A, 2023. 120(8): p. e2220415120.
- Guarner J, Johnson BJ, Paddock CD, Shieh WJ, Goldsmith CS, Reynolds MG, Damon IK, Regnery RL, Zaki SR; Veterinary Monkeypox Virus Working Group. Monkeypox transmission and pathogenesis in prairie dogs. Emerg Infect Dis. 2004 Mar;10(3):426-31. doi: 10.3201/eid1003.030878. PMID: 15109408; PMCID: PMC3322777.
- Parker, S. and R.M. Buller, A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol, 2013. 8(2): p. 129-157.
- Ladnyj, I.D., P. Ziegler, and E. Kima, A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ, 1972. 46(5): p. 593-7.
- Hutin, Y.J., et al., Outbreak of human monkeypox, Democratic Republic of Congo, 1996 to 1997. Emerg Infect Dis, 2001. 7(3): p. 434-8.
- Sergeev, A.A., et al., The Possibility of Using the ICR Mouse as an Animal Model to Assess Antimonkeypox Drug Efficacy. Transbound Emerg Dis, 2016. 63(5): p. e419-30.
- JC., H. and K. RA., 2 - Virus isolation and quantitation A2. Virology methods manual . , ed. B.W. Mahy and K. HO. 1996, London,: Academic Press. 25-46.
- Aid M, Sciacca M, McMahan K, Hope D, Liu J, Jacob-Dolan C, Powers O, Barrett J, Wu C, Mutoni A, Murdza T, Richter H, Velasco J, Teow E, Boursiquot M, Cook A, Orekov T, Hamilton M, Pessaint L, Ryan A, Hayes T, Martinot AJ, Seaman MS, Lewis MG, Andersen H, Barouch DH. Mpox infection protects against re-challenge in rhesus macaques. Cell. 2023 Oct 12;186(21):4652-4661.e13. doi: 10.1016/j.cell.2023.08.023. Epub 2023 Sep 20. PMID: 37734373; PMCID: PMC10591870.
- Li, Y., et al., Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J Virol Methods, 2010. 169(1): p. 223-7.
- Test Procedure: Monkeypox virus Generic Real-Time PCR Test. 2022
- Li, Y., et al., Detection of monkeypox virus with real-time PCR assays. J Clin Virol, 2006. 36(3): p. 194-203.
- Test Procedure: Non-variola Orthopoxvirus Generic Real-Time PCR Test. 2022.
- Doty, J.B., et al., Assessing Monkeypox Virus Prevalence in Small Mammals at the Human-Animal Interface in the Democratic Republic of the Congo. Viruses, 2017. 9(10).
- Hubert, M., et al., Complement-dependent mpox-virus-neutralizing antibodies in infected and vaccinated individuals. Cell Host Microbe, 2023. 31(6): p. 937-948.e4.
- Davies, D.H., et al., Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J Virol, 2005. 79(18): p. 11724-33.
- Zhou, B., et al., Two long-lasting human monoclonal antibodies cross-react with monkeypox virus A35 antigen. Cell Discov, 2023. 9(1): p. 50.
- Lansiaux, E., et al., The virology of human monkeypox virus (hMPXV): A brief overview. Virus Res, 2022. 322: p. 198932.
- Soekawa, M., et al., Electron-microscopical observations on the development of vaccinia, cowpox and monkeypox viruses in pig skin. Zentralbl Bakteriol Orig A, 1977. 237(4): p. 425-43.
- Meister, T.L., et al., Stability and inactivation of monkeypox virus on inanimate surfaces. J Infect Dis, 2023.
- Hooper, J.W., et al., Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. J Virol, 2004. 78(9): p. 4433-43.
- Stittelaar, K.J., et al., Modified vaccinia virus Ankara protects macaques against respiratory challenge with monkeypox virus. J Virol, 2005. 79(12): p. 7845-51.
- Nalca, A., et al., Experimental infection of cynomolgus macaques (Macaca fascicularis) with aerosolized monkeypox virus. PLoS One, 2010. 5(9).
- Hutson, C.L., et al., Monkeypox disease transmission in an experimental setting: prairie dog animal model. PLoS One, 2011. 6(12): p. e28295.
- Johnson, R.F., et al., Comparative analysis of monkeypox virus infection of cynomolgus macaques by the intravenous or intrabronchial inoculation route. J Virol, 2011. 85(5): p. 2112-25.
- Hutson, C.L., et al., Comparison of Monkeypox Virus Clade Kinetics and Pathology within the Prairie Dog Animal Model Using a Serial Sacrifice Study Design. Biomed Res Int, 2015. 2015: p. 965710.
- Kretzschmar, M., et al., Ring vaccination and smallpox control. Emerg Infect Dis, 2004. 10(5): p. 832-41.
- Ward, T., et al., Transmission dynamics of monkeypox in the United Kingdom: contact tracing study. BMJ, 2022. 379: p. e073153.
- Deschambault, Y., et al., Experimental Infection of North American Deer Mice with Clade I and II Monkeypox Virus Isolates. Emerg Infect Dis, 2023. 29(4): p. 858-860.
- Khodakevich, L., Z. Jezek, and D. Messinger, Monkeypox virus: ecology and public health significance. Bull World Health Organ, 1988. 66(6): p. 747-52.
- Nolen, L.D., et al., Introduction of Monkeypox into a Community and Household: Risk Factors and Zoonotic Reservoirs in the Democratic Republic of the Congo. Am J Trop Med Hyg, 2015. 93(2): p. 410-5.
- Breman, J.G., J. Bernadou, and J.H. Nakano, Poxvirus in West African nonhuman primates: serological survey results. Bull World Health Organ, 1977. 55(5): p. 605-12.

