ABSTRACT
Interleukin (IL)-37 is an anti-inflammatory cytokine that has recently been proposed to have an etio-pathogenic role in systemic lupus erythematosus (SLE). Two exon variants, rs3811046 G/T and rs3811047 A/G, of the IL37 gene have also been associated with susceptibility to some autoimmune disorders but have not been investigated in SLE. Therefore, this case-control study was conducted on 120 women with SLE and 120 healthy control women to explore the association between both variants and susceptibility to SLE. In addition, serum IL-37 levels were also determined. The TaqMan allelic discrimination method was used to genotype rs3811046 and rs3811047, while an ELISA kit was used to quantify IL-37 levels. IL-37 levels were significantly decreased in SLE patients compared to controls. Mutant alleles (T and G, respectively) as well as corresponding homozygous genotypes (TT and GG, respectively) of rs3811046 and rs3811047 were associated with a reduced risk of SLE. Haplotype analysis (in the order rs3811046–rs3811047) demonstrated that the T-A and G-A haplotypes were associated with an increased risk of SLE (2.53-fold and 2.45-fold, respectively). In conclusion, IL37 variants rs3811046 and rs3811047 were associated with reduced susceptibility to SLE among Iraqi women but were not associated with risk of active disease or lupus nephritis.
Introduction
Systemic lupus erythematosus (ЅLЕ) is worldwide disease with an estimated global incidence of 5.14 per 100,000 population per year. The incidence varies widely between women and men (8.82 versus 1.53 per 100,000 population per year), as well as across ethnic and geographic regions [Citation1]. SLE is a systemic inflammatory autoimmune disorder with heterogeneous clinical manifestations including mild cutaneous and mucosal manifestations that can progress to affect multiple organs, especially joints and kidneys [Citation2]. The etiology and pathophysiology of SLE have not been precisely determined but there is compelling evidence suggesting that the interaction between genetic, endocrine and environmental factors can cause loss of immune tolerance to self-antigens associated with dysregulated functions of the innate and adaptive immune systems [Citation3]. Several immunopathogenic pathways have been shown to participate in the initiation and progression of ЅLЕ and ultimately lead to the formation of pathogenic autoantibodies, in particular anti-double stranded DNА antibodies (anti‐dsDNA). This is accompanied by dysregulated levels of complement proteins, especially C3 and C4 [Citation4]. Equally important, studies have also revealed that cytokines, low-molecular-weight proteins involved in coordinating and mediating inflammatory and autoimmune reactions, play a pathophysiological role in SLE. Some cytokines have been linked to the etiology of SLE and may therefore be useful biomarkers in prognosticating the disease and monitoring its activity [Citation5]. Moreover, it has recently been shown that some cytokines, such as interferon (IFN)-γ, interleukin (IL)-17, and IL-10, may influence the production of autoantibodies involved in the pathogenesis of SLE along with their association with clinical manifestations of the disease [Citation6]. Therefore, cytokines may be considered potential biomarkers to evaluate the activity, severity, and manifestations of SLE and may also predict disease remission especially when their genetic variants (i.e. single nucleotide polymorphisms; SNPs) are evaluated for association with disease risk [Citation7].
One cytokine that has attracted interest in the field of autoimmunity in general and SLE in particular is IL-37. It is a cytokine that has been assigned to the ΙL-1 cytokine family in recent years, and its pathogenic potential has been suggested as a biomarker associated with an increased risk of autoimmune and inflammatory diseases [Citation8,Citation9]. ΙL-37 (formerly known as IL-1 family member 7; IL-1F7) is a cytokine with anti-inflammatory effects that acts on monocytes/macrophages and epithelial cells and suppresses these cells from releasing cytokines with pro-inflammatory function such as ΙL-1α, IL-1β, and tumor necrosis factor (TNF)-α [Citation10]. Regarding inflammatory and autoimmune diseases, most studies have shown dysregulation of IL-37 production and its anti-inflammatory effects have been reported in a number of these diseases, including rheumatoid arthritis, ankylosing spondylitis, Graves’ disease, and inflammatory bowel disease. Furthermore, dysregulated IL-37 levels have been linked to the severity and activity of these diseases [Citation8,Citation11]. With respect to SLE, It has recently been demonstrated that ΙL-37 may signature the etiopathogenesis of the disease because IL-37 showed dysregulated levels in patients with SLE. In addition, abnormally expressed IL-37 has been linked to disease activity and correlated with pro-inflammatory mediators, such as IL-6, IL-18, and IFN-γ. However, the association of IL-37 with clinical manifestations of SLE, such as lupus nephritis (LN), arthritis, and cutaneous manifestations, has not been well explored [Citation9,Citation12–14].
IL-37 is encoded by a gene located on human chromosome 2 at position 2q14.1 and consists of seven exons and spans 3617 kilo-base pairs (kb) (www.ncbi.nlm.nih.gov/gene/27178). The IL37 gene contains several naturally occurring SNPs, and case-control studies have linked some of these genetic variants with a predisposition to developing several infectious diseases, autoimmune diseases, and inflammatory diseases, including SLE [Citation9,Citation15]. Among these variants, two exon SNPs with a missense mutation, rs3811046 and rs3811047, have recently been suggested to be associated with the risk of developing certain inflammatory and autoimmune diseases, such as periodontitis, ankylosing spondylitis, rheumatoid arthritis, autoimmune thyroid diseases, and Behçet’s disease [Citation16–18]. In relation to SLE, these two SNPs of the IL37 gene have not been investigated. Therefore, a case-control study was conducted to evaluate whether IL37 SNPs rs3811046 and rs3811047 were associated with susceptibility to SLE among Iraqi women. Meanwhile, the relationship between these polymorphisms and disease activity and other clinical parameters has been explored. In addition, the effect of the SNPs on serum IL-37 was analyzed.
Materials and methods
Ethical statement
The Ethics Committee at the Department of Biotechnology (College of Science, University of Baghdad) and the Baghdad Medical City Complex (Iraqi Ministry of Health and Environment) approved the study protocol (Reference No.: CSEC/1121/0078 on 20 November 2021 and 2084 on 16 January 2022, respectively). All participants were informed of the study objectives and provided written consent.
Participants
This case-control study was performed on 120 women diagnosed with SLE and a similar number of age-matched healthy control women (HCW) (mean ± standard deviation (SD) = 34.0 ± 10.2 and 35.9 ± 9.6 years, respectively). Patients were attending the SLE unit at the Nephrology and Renal Transplantation Centre (Baghdad Medical City Complex) during January-November 2022. Diagnosis followed the American Rheumatology College criteria for SLE [Citation19]. Only females at the age of 18 years and above and followed the SLE diagnostic criteria were included. Patients with chronic diseases, including diabetes, cardiovascular disease and cancer, and pregnant women were excluded. The SLE Disease Activity Index (SLEDAI) was used to evaluate SLE activity. A score < 10 indicates mild/moderate disease activity while a score ≥ 10 indicates active disease [Citation20]. Diagnosis of LN was based on 24-h urinalysis (proteinuria greater than 0.5 gram per day). Besides, blood urea nitrogen (BUN) and serum creatinine (SCR) were determined [Citation21]. All patients were on treatment including prednisolone (10 mg/day) plus mycophenolate mofetil (2 mg/day) or tacrolimus (1 mg/day). A questioner was filled out and included baseline characteristic data used to describe patients with SLE. These data were collected from the hospital registry and included age of patients, duration of SLE, body mass index (BMI), SLEDAI, medication, BUN, SCR, hemoglobin (Hb) level, platelet count, white blood cell (WBϹ) count, C3, C4, anti-nuclear antibody (ANA) and anti-dsDNA antibodies. The HCW group included blood donors (National Blood Transfusion Centre, Baghdad) and health workers who were in good health and free of chronic or infectious diseases.
IL-37 immunoassay
IL-37 levels in participants’ serum were measured using a commercially available enzyme-linked immunosorbent assay (ELISA) kit, which was a product of MyBioSource (USA) under catalog number MBS165041. The analytical technique recommended by the manufacturer was followed and testing was performed using an ELISA microplate reader (HumaReader HS, Human Diagnostics, Germany). The detection range of the kit was 0.5–200 ng/L with a sensitivity of 0.25 ng/L.
DNA isolation and SNP detection
The EasyPure Blood gDNA kit was used to isolate genomic DNA from whole blood following the manufacturer’s protocol (Transgen Biotech, China). A NanoDrop spectrophotometer (Quawell, USA) was used to assess DNA concentration and purity.
The TaqMan allelic discrimination method was used to genotype the IL37 SNPs rs3811046 G/T and rs3811047 A/G. Both SNPs are exon 2 missense variants close to each other (31 nucleotides apart) and their chromosomal locations are chromosome 2:112913801 and chromosome 2:112913833, respectively (www.ensembl.org). The method was based on the principles of real-time polymerase chain reaction (RT-PCR) testing. Two primers (forward and reverse) and two probes (for detection of wild-type and mutant alleles) were designed for each SNP (Primer3Plus software; www.primer3plus.com) based on their National Center for Biotechnology Information (NCBI) reference sequences. For rs3811046, primers were 5’- CATATGCTAACCTCACTGCGT-3“ (forward) and 5”-AATTCATGGTGGGGAGGCTT-3“ (reverse) and probes were FAM-TGGAAGCCCCCTGGAAC-BHQ and JOE-CTGTAAGCCCCCTGGAACC-BHQ. For rs3811047, primers were 5”-CATATGCTAACCTCACTGCGT-3“ (forward) and 5”-CCCATCACCTCACCCCGA-3’ (reverse) and probes were CY3-CCCACCATGAATTTTGTTCACAC-BHQ and CY5- CCGCCATGAATTTTGTTCACAC-BHQ. The efficiency and specificity of primers and probes were tested online using in-silico PCR analysis (genome.ucsc.edu/cgi-bin/hgPcr). Primers and probes were provided by Alpha DNA (Canada).
The RT-PCR mix (total volume = 20 µL) consisted of 4 µL super mix (PerfectStart II Probe qPCR SuperMix UDG, TransgenBiotech, China), forward and reverse primers (1.3 µL each), probes 1 and 2 (0.8 µL each), 4.5 µL DNA, and 7.3 µL nuclease-free water. The RT-PCR protocol was 95°C (1 min), followed by 36 cycles of 95°C (15 sec), 60°C (1 min), and 72°C (1 min) for rs3811046 and 95°C (10 min), followed by 40 cycles of 95°C (5 sec), 58°C (1 min), and 72°C (30 sec) for rs3811047. MxPro 3005P qPCR System was used to perform the RT-PCR protocol and MxPro software (Stratagene, USA) was used to interpret data.
Statistical analysis
Most continuous variables did not follow a normal distribution (Kolmogorov-Smirnov and the Shapiro-Wilk tests) and were therefore presented by the median and interquartile range (IQR: 25–75%) and significance was detected using the Mann-Whitney or Kruskal-Wallis test. Number and percentage were used to describe categorical variables and two-tailed Fisher’s exact test was used to test significance. Hardy-Weinberg equilibrium (HWE) analysis was performed using Pearson Chi-Square test. The area under the curve (AUC), 95% confidence interval (CI), cutoff point, sensitivity, and specificity were calculated using receiver operating characteristic (ROC) curve analysis. Odds ratio (OR) and 95% CI were estimated using multinomial logistic regression analysis. A probability (p) < 0.05 was considered statistically significant. Bonferroni correction was applied to correct the p-value (pc). Statistical analysis was performed with IBM SPSS Statistics 25.0 (Armonk, NY: IBM Corp.) and figures were plotted with GraphPad Prism version 9.2.0 (San Diego, CA, USA). SHEsis software was used to estimate linkage disequilibrium (LD) and to carry out haplotype analysis [Citation22].
Results
Baseline data
Median age (IQR) of SLE women was 32.0 (26.0–42.5) years, which was not significantly different from that of HCW (34.0 [28.0–41.5] years; р = 0.143). In terms of age groups (≤40 and >40 year), SLE patients and HCW also did not show a significant difference (p = 0.318). Disease duration was 4.0 (2.0–6.0) years and was divided into four groups: 1–2 (30.0%), 3–4 (29.2%), 5–6 (16.7%), and > 6 (24.2%) years. The BMI of the patients was 28.6 (25.0–31.7) kg/m2. In fact, most patients were overweight/obese (76.7%). According to SLEDAI, SLE was grouped into mild/moderate disease (85.8%) and active disease (14.2%). It was also found that 23.3% of SLE patients had LN (). Laboratory data for SLE patients are also given in .
Table 1. Baseline clinical and laboratory data for SLE patients and healthy control women.
IL-37 levels
IL-37 showed significantly lower levels in SLE patients than in HCW (34.4 [28.0–45.8] vs. 54.1 [39.0–68.3] ng/L; p < 0.001). ROC curve analysis demonstrated the performance of IL-37 in distinguishing between patients and controls (AUC = 0.785; 95% CI = 0.782–0.841; р < 0.001). At a cutoff point of 40.6 ng/L (YI = 0.39), the sensitivity and specificity of IL-37 were 70.8 and 68.3%, respectively (). When IL-37 levels were stratified by age (≤40 and >40 years), disease duration (1–2, 3–4, 5–6, and >6 years), BMI (normal weight and overweight/obese), SLEDAI (mild/moderate and active), and LN (present and absent), no significant difference was found in each stratification, except SLEDAI. Patients with active disease were characterized by significantly higher levels of IL-37 compared to patients with mild/moderate disease (40.1 [32.2–52.4] vs. 34.0 [26.1–43.6] ng/L; р = 0.049) ().
Figure 1. [a] Box-whisker plot of serum IL-37 levels in SLE patients and healthy control women (HCW). The horizontal line inside the box indicates the median. Whiskers indicate interquartile range (IQR). Black circles indicate outliers. Mann-Whitney U test was used to assess significant difference (***p < 0.001). [b] Receiver operating characteristic (ROC) curve analysis of IL-37 in SLE patients versus HCW. AUC: area under the curve; CI: confidence interval; p: probability.
![Figure 1. [a] Box-whisker plot of serum IL-37 levels in SLE patients and healthy control women (HCW). The horizontal line inside the box indicates the median. Whiskers indicate interquartile range (IQR). Black circles indicate outliers. Mann-Whitney U test was used to assess significant difference (***p < 0.001). [b] Receiver operating characteristic (ROC) curve analysis of IL-37 in SLE patients versus HCW. AUC: area under the curve; CI: confidence interval; p: probability.](/cms/asset/5558771b-15f2-4ea4-9635-02a23ec0e997/teba_a_2336702_f0001_b.gif)
Table 2. Serum interleukin-37 levels stratified by characteristics of SLE patients.
IL37 SNPs
Two SNPs in exon 2 of the IL37 gene were investigated; rs3811046 G/T and rs3811047 A/G. The TaqMan allelic discrimination method was used to genotype the two SNPs. This method revealed that rs3811046 had three genotypes, which were GG, GT, and TT. The rs3811047 was also characterized with three genotypes, which were AA, AG, and GG. Genotype frequencies of rs3811046 and rs3811047 were consistent with HWE in the HCW sample (p = 0.174 and 0.163, respectively), while genotype frequencies of both ЅΝРs were significantly deviated from the HWE in SLE patients (p = 0.039 and 0.001, respectively) ().
Table 3. Association analysis of IL37 gene SNPs rs3811046 and rs3811047 with SLE risk.
A comparison between SLE patients and HCW revealed that T allele of гѕ3811046 exhibited a significantly decreased frequency in patients compared with HCW (55.8 vs. 66.2%; p = 0.025). This allele was found to be associated with a reduced risk of SLE (OR = 0.64; 95% CI = 0.45–0.93). Similarly, the TT genotype exhibited a significantly decreased frequency in patients compared with HCW (35.8 vs. 46.6%; p = 0.033). This genotype was also associated with a decreased risk of SLE (OR = 0.45; 95% CI = 0.22–0.92). However, the significance in both cases was not maintained when Bonferroni correction was applied (pc = 0.075 and 0.099, respectively) ().
Regarding SNP гѕ3811047, allele G (51.2 vs. 77.1%; p < 0.001) and genotype GG (35.8 vs. 61.7%; p < 0.001) exhibited significantly decreased frequencies in SLE patients compared with HCW, and were associated with a reduced risk of SLE (OR = 0.31 and 0.13; 95% CI: 0.21–0.46 and 0.06–0.29, respectively). For genotype AG, although it showed a similar frequency in patients and controls (30.8%), it was linked to a significantly lower risk of SLE versus AA genotype (OR = 0.22; 95% CI = 0.10–0.52; p < 0.001). These differences remained significant after applying Bonferroni correction (pc < 0.001) ().
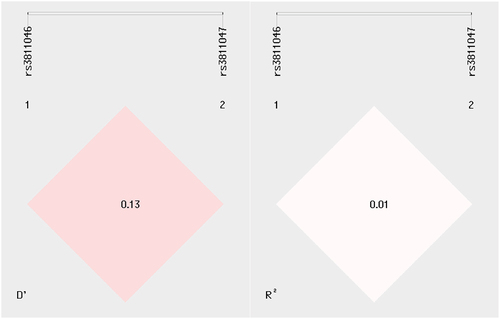
LD analysis revealed that rs3811046 and rs3811047 were in very weak LD as indicated by the estimated D’ (0.13) and R2 (0.01) values (). Haplotype analysis (in the order rs3811046–rs3811047) showed that frequency of haplotype T-A was significantly higher in SLE patients compared with HCW (25.8 vs. 12.0%; p < 0.001). Haplotype G-A also demonstrated a significantly increased frequency in patients (22.9 vs. 10.8; p < 0.001). The deviation of both haplotypes was associated with an increased risk of SLE (OR = 2.53 [95% CI = 1.56–4.11] for T-A haplotype and 2.45 [95% CI = 1.47–4.06] for G-A haplotype). On the contrary, haplotype T-G was associated with a significantly reduced risk of SLE (30.0 vs. 54.1; OR = 0.36; 95% CI = 0.25–0.53; p < 0.001). In all comparisons, the p-value remained significant after applying Bonferroni correction ().
Association of IL37 SNPs with SLEDAI and LN
The allele and genotype frequencies of rs3811046 and rs3811047 were examined in SLE patients after classification by SLEDAI (mild/moderate and active) and LN (present and absent) groups. Results revealed that neither allele nor genotype frequencies of these SNPs showed significant differences between patients with respect to both classifications ().
Table 4. Relationship of IL37 gene variants rs3811046 and rs3811047 to disease activity and lupus nephritis.
Impact of IL37 SNP genotypes on IL-37 levels
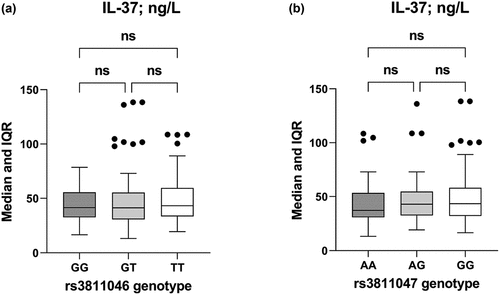
When IL-37 levels were stratified by rs3811046 and rs3811047 genotypes, the levels were almost approximated in the three genotypes of each ЅΝР (i.e. no significant difference) ().
Figure 3. Box-whisker plot of serum IL-37 levels in SLE patients plus healthy control women stratified by genotypes of IL37 SNPs rs3811046 and rs3811047. The horizontal line inside the box indicates the median. Whiskers indicate interquartile range (IQR). Black circles indicate outliers. Mann-Whitney U test was used to assess significant difference (ns: not significant).
Discussion
In this study, serum levels and two SNPs of IL-37 were studied in women with SLE and results were compared with HCW. IL-37 showed down-regulated levels in SLE patients and achieved reliable performance in distinguishing between patients and controls (AUC = 0.785). IL-37 levels were not associated with age, disease duration, BMI, or LN, but were significantly elevated in patients with active disease compared with patients with mild/moderate disease activity. IL-37 is an IL-1 family cytokine with anti-inflammatory functions, and its serum/plasma levels have been shown to be dysregulated in most inflammatory and autoimmune conditions [Citation8]. With an increase in pro-inflammatory cytokines, anti-inflammatory cytokines are expected to modulate inflammatory reactions by counteracting the effects of pro-inflammatory cytokines. Therefore, their levels should be raised. Otherwise, if anti-inflammatory cytokines are unable to mediate these functions, inflammation arises and progresses [Citation23]. Indeed, over-expression of IL-37 has been found to be associated with up-regulated expression of some cytokines with pro-inflammatory potential such as IL-6. In addition, abnormal concentrations of IL-37 have been indicated in a number of autoimmune diseases, as well as inflammatory disorders. For example, IL-37 showed higher levels in patients with Graves’ disease, rheumatoid arthritis, or ankylosing spondylitis, while patients with inflammatory bowel disease showed down-regulated levels. Therefore, IL-37 has been linked to the pathogenesis of these diseases [Citation24,Citation25]. Regarding SLE, there is almost unanimous agreement among studies that IL-37 levels are up-regulated in patients with the disease [Citation9,Citation12–14]. The results of the current study were not consistent with these studies. On the contrary, it showed decreased levels of IL-37 in patients with SLE. This discrepancy could be attributed to the immune status of the present patients who were all taking immunosuppressive medications to control their autoimmune and inflammatory condition. Supporting evidence indicated that SLE patients who received prednisone treatment for 14 days showed a significant decrease in IL-37 levels [Citation26]. However, the present study revealed that IL-37 was associated with SLE activity, and that its levels were significantly increased in patients with active disease compared with those with mild/moderate activity. In agreement with our observation, a recent meta-analysis of 14 publications demonstrated a positive correlation between circulatory IL-37 levels and SLE activity [Citation27]. Therefore, IL-37 may be considered a risk factor associated with SLE activity despite the use of immunosuppressive drugs.
The main interest of the current study was to evaluate the significance of two IL37 SNPs, rs3811046 and rs3811047, in SLE susceptibility as these SNPs have shown association with a number of autoimmune and inflammatory diseases but have not been studied in SLE. The results indicated that alleles (T and G) and genotypes (TT and GG) of both SNPs (rs3811046 and rs3811047, respectively), especially rs3811047, were associated with a lower risk of SLE among Iraqi women. The most informative association profile was identified when performing haplotype estimation of SNPs. The T-A and G-A haplotypes (rs3811046–rs3811047) were respectively associated with a 2.53-fold and 2.45-fold increased risk of SLE, while a decreased risk was associated with the T-G haplotype. These findings suggest that both genetic variants of the IL37 gene may influence susceptibility to SLE. SNPs rs3811046 and rs3811047 are two missense variants of the IL37 gene that are close to each other and separated by only 31 nitrogenous bases. Missense mutations (G > T for rs3811046 and A > G for rs3811047) result in the replacement of glycine with valine and threonine with alanine (amino acids), respectively. These molecular consequences may be considered risk factors for the development of some infectious, inflammatory, and autoimmune diseases [Citation15].
Regarding rs3811046, some studies reported no association between this ЅΝР and Hashimoto’s thyroiditis, primary open-angle glaucoma, tuberculosis, and rheumatoid arthritis [Citation28–31]. Other studies, however, have linked rs3811046 to susceptibility to other infectious, inflammatory, and autoimmune diseases such as periodontitis, Graves’ disease and COVID-19 [Citation15,Citation18,Citation28]. In the current study, the evidence of association between rs3811046 and SLE was weak because significance was not maintained after p-value correction for multiple comparisons. In the case of SNP 3,811,047, stronger evidence from this study indicated that both the G allele and the GG genotype were considered factors that could be involved in reducing the risk of SLE and the association was highly significant. In other studies 3,811,047 has also been associated with susceptibility to Behcet’s disease, ankylosing spondylitis, rheumatoid arthritis and peptic ulcer due to infection with H. pylori but the evidence was not overwhelming [Citation16,Citation17,Citation32,Citation33]. Although the results of these studies have not been confirmed or replicated, together they suggest the significance of rs3811047 in susceptibility to autoimmune diseases including SLE as demonstrated in this study. However, alleles and genotypes of both IL37 SNPs showed no significant association with SLE activity or LN. In addition, there was no significant effect of these genetic variants on serum levels of IL-37.
All patients in the current study were receiving immunosuppressive medications. The results for IL-37 levels should therefore be interpreted with caution, and in fact this was an important limitation of the study. Including newly diagnosed cases and untreated patients, the role of IL-37 in the etio-pathogenesis of SLE will be better evaluated. In addition, the number of patients with active disease and those who developed LN was relatively small this can be considered an additional limitation in the current study.
Conclusions
IL37 variants rs3811046 and rs3811047 were associated with reduced susceptibility to SLE among Iraqi women but were not associated with risk of active disease or LN. Furthermore, rs3811046 and rs3811047 genotypes had no effect on serum IL-37 levels.
Acknowledgments
The authors appreciate the assistance provided by the medical staff at the Center for Nephrology and Kidney Transplantation (Baghdad Medical City Complex) in conducting this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Tian J, Zhang D, Yao X, et al. Global epidemiology of systemic lupus erythematosus: a comprehensive systematic analysis and modelling study. Ann Rheum Dis. 2023;82:351–356. doi: 10.1136/ard-2022-223035
- Sticherling M, Kuhn A. Lupus erythematosus. Braun-Falco’s Dermatology. Berlin Heidelberg: Springer; 2022. p. 923–939. doi: 10.1007/978-3-662-63709-8_54
- Ameer MA, Chaudhry H, Mushtaq J, et al. An overview of systemic lupus erythematosus (SLE) pathogenesis, classification, and management. Cureus. 2022;14:e30330. doi: 10.7759/cureus.30330
- Damoiseaux J, van Beers J. Autoantibodies to dsDNA in the diagnosis, classification and follow-up of patients with systemic lupus erythematosus. J Transl Autoimmun. 2023;6:100191. doi: 10.1016/j.jtauto.2023.100191
- Rincón-Delgado KL, Tovar-Sánchez C, Fernández-Ávila DG, et al. Role of cytokines in the pathophysiology of systemic lupus erythematosus. Rev Colomb Reumatol. 2021;28:144–155. doi: 10.1016/j.rcreu.2021.05.018
- Alduraibi FK, Sullivan KA, Chatham WW, et al. Interrelation of T cell cytokines and autoantibodies in systemic lupus erythematosus: a cross-sectional study. Clin Immunol. 2023;247:109239. doi: 10.1016/j.clim.2023.109239
- Alduraibi FK, Tsokos GC. Lupus nephritis biomarkers: a critical review. Int J Mol Sci. 2024;25:805. doi: 10.3390/ijms25020805
- Zeng H, Zhou K, Ye Z. Biology of interleukin‑37 and its role in autoimmune diseases (review). Exp Ther Med. 2022;24:495. doi: 10.3892/etm.2022.11422
- Wu Q, Zhou J, Yuan ZC, et al. Association between IL-37 and systemic lupus erythematosus risk. Immunol Invest. 2022;51:727–738. doi: 10.1080/08820139.2020.1869254
- Yan X, Xie B, Wu G, et al. Interleukin-37: the effect of anti-inflammatory response in human coronary artery endothelial cells. Mediators Inflamm. 2019;2019:2650590. doi: 10.1155/2019/2650590
- Su Z, Tao X. Current understanding of IL-37 in human health and disease. Front Immunol. 2021;12:696605. doi: 10.3389/fimmu.2021.696605
- Wu GC, Li HM, Wang JB, et al. Elevated plasma interleukin-37 levels in systemic lupus erythematosus patients. Lupus. 2016;25:1377–1380. doi: 10.1177/0961203316646462
- Godsell J, Rudloff I, Kandane-Rathnayake R, et al. Clinical associations of IL-10 and IL-37 in systemic lupus erythematosus. Sci Rep. 2016;6:1–10. doi: 10.1038/srep34604
- Ye L, Ji L, Wen Z, et al. IL-37 inhibits the production of inflammatory cytokines in peripheral blood mononuclear cells of patients with systemic lupus erythematosus: its correlation with disease activity. J Transl Med. 2014;12:69. doi: 10.1186/1479-5876-12-69
- Ahmed AA, Ad’hiah AH. Interleukin-37 gene polymorphism and susceptibility to coronavirus disease 19 among Iraqi patients. Meta Gene. 2022;31:100989. doi: 10.1016/j.mgene.2021.100989
- Özgüçlü S, Duman T, Ateş FSÖ, et al. Serum interleukin-37 level and interleukin-37 gene polymorphism in patients with behçet disease. Clin Rheumatol. 2019;38:495–502. doi: 10.1007/s10067-018-4288-7
- Lin XY, Guo XJ, He YZ, et al. Association between interleukin 37 (rs3811047) polymorphism and multiple autoimmune diseases in a Chinese population. Med (United States). 2018;97(15):e0386. doi: 10.1097/MD.0000000000010386
- Cirelli T, Nepomuceno R, Orrico SRP, et al. Validation in a Brazilian population of gene markers of periodontitis previously investigated by GWAS and bioinformatic studies. J Periodontol. 2021;92:689–703. doi: 10.1002/JPER.20-0126
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928
- Koelmeyer R, Nim HT, Nikpour M, et al. High disease activity status suggests more severe disease and damage accrual in systemic lupus erythematosus. Lupus Sci Med. 2020;7:e000372. doi: 10.1136/lupus-2019-000372
- Siedner MJ, Gelber AC, Rovin BH, et al. Diagnostic accuracy study of urine dipstick in relation to 24-hour measurement as a screening tool for proteinuria in lupus nephritis. J Rheumatol. 2008;35(1):84–90.
- Shi YY, He L. Shesis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272
- Wautier JL, Wautier MP. Pro- and anti-inflammatory prostaglandins and cytokines in Humans: a mini review. Int J Mol Sci. 2023;24:9647. doi: 10.3390/ijms24119647
- Jia H, Liu J, Han B. Reviews of interleukin-37: functions, receptors, and roles in diseases. Biomed Res Int. 2018;2018:3058640. doi: 10.1155/2018/3058640
- Wang L, Quan Y, Yue Y, et al. Interleukin-37: a crucial cytokine with multiple roles in disease and potentially clinical therapy (review). Oncol Lett. 2018;15:4711. doi: 10.3892/OL.2018.7982
- Song L, Qiu F, Fan Y, et al. Glucocorticoid regulates interleukin-37 in systemic lupus erythematosus. J Clin Immunol. 2013;33:111–117. doi: 10.1007/s10875-012-9791-z
- Lee YH, Song GG. Circulating interleukin-37 levels in Rheumatoid Arthritis and Systemic lupus erythematosus and their correlations with disease activity: a meta-analysis. J Rheum Dis. 2020;27:152–158. doi: 10.4078/jrd.2020.27.3.152
- Yan N, Meng S, Song RH, et al. Polymorphism of IL37 gene as a protective factor for autoimmune thyroid disease. J Mol Endocrinol. 2015;55:209–218. doi: 10.1530/JME-15-0144
- Zhang XY, Zuo Y, Li C, et al. IL1F7 gene polymorphism is not associated with rheumatoid arthritis susceptibility in the northern Chinese han population: a case-control study. Chin Med J (Engl). 2018;131:171–179. doi: 10.4103/0366-6999.222340
- Allam G, Mohamed IAA, Alswat KA, et al. Association of IL-37 gene polymorphisms with susceptibility to tuberculosis in Saudi subjects. Microbiol Immunol. 2016;60:778–786. doi: 10.1111/1348-0421.12444
- Mookherjee S, Banerjee D, Chakraborty S, et al. Evaluation of the IL1 gene cluster single nucleotide polymorphisms in primary open-angle glaucoma pathogenesis. Genet Test Mol Biomarkers. 2016;20:633–636. doi: 10.1089/gtmb.2015.0344
- Davarpanah E, Jafarzadeh A, Nemati M, et al. Circulating concentration of interleukin-37 in helicobacter pylori-infected patients with peptic ulcer: its association with IL-37 related gene polymorphisms and bacterial virulence factor CagA. Cytokine. 2020;126:154928. doi: 10.1016/j.cyto.2019.154928
- Tan H, Deng B, Yu H, et al. Genetic analysis of innate immunity in Behcet’s disease identifies an association with IL-37 and IL-18RAP. Sci Rep. 2016;6:35802. doi: 10.1038/srep35802