?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Agricultural modernization urgently requires precise control of pear tree leaf diseases, with accurate identification and segmentation of disease spots becoming crucial aspects for ensuring crop health. Addressing potential issues such as missed segmentation, missegmentation, and low segmentation accuracy in the traditional DeepLabv3+ model for this task, this study proposes an innovative approach based on an improved DeepLabv3+ network model. To enhance computational efficiency, MobileNetV2 is introduced as the backbone network. This reduces the model’s computational load and significantly improves segmentation speed, making it more suitable for real-time applications. Secondly, after the ASPP (Atrous Spatial Pyramid Pooling) convolution, the Squeeze-and-Excitation (SE) attention mechanism is introduced, integrating the features of pear tree leaf diseases to make the network focus more on the critical components of disease spots, thereby enhancing segmentation accuracy. Finally, the loss function is optimized by employing a linear combination of the cross-entropy loss function and Dice loss, encompassing comprehensive overall image loss and accurate loss calculation for the target area. Experimental validation demonstrates a significant improvement in the enhanced DeepLabv3+ across metrics such as MIoU, mPA, mPr, and mRecall, reaching 86.32%, 88.97%, 91.10%, and 88.97%, respectively. The improved model excels in pear tree leaf disease segmentation compared to traditional methods like DeepLabv3+, SegNet, FCN, and PSPNet. This confirms the enhanced model’s outstanding performance and generalization ability in segmenting pear tree leaf lesions, highlighting its potential value in practical applications.
REVIEWING EDITOR:
1. Introduction
Pear trees, a crucial fruit crop, are susceptible to various fungi, bacteria, and viruses during their growth, leading to leaf diseases (Yanjun et al., Citation2021). Failure to promptly identify and control the spread of leaf diseases in pear trees can ultimately result in a decline in yield and quality, affecting the economic benefits of farmers (Wang, Citation2020). With the increasing demand for managing leaf diseases, traditional disease detection methods often rely on manual inspection, which is time-consuming and prone to subjective errors. Using efficient and accurate leaf disease segmentation methods based on deep learning becomes crucial.
Image segmentation is a crucial component of image analysis and understanding in computer vision, particularly in identifying and extracting relevant features for plant leaf disease segmentation. The segmentation results provide a favorable basis for calculating leaf area and disease recognition. Traditional leaf segmentation methods encompass threshold-based approaches (Lincai et al., Citation2022), edge detection techniques (Donghui et al., Citation2016; Semma et al., Citation2021), region-based (Zhongyuan et al., Citation2004), and clustering-based algorithms (Ji-Feng, Citation2022). Traditional segmentation techniques are well-established for most segmentation tasks. However, they face time consumption issues and are unsuitable for real-time detection in fast-paced scenarios. Conventional methods may struggle to identify the type and severity of diseases accurately. In contrast, deep learning models can learn complex image features, achieving precise segmentation and quantitative Analysis of different conditions (Wenbo et al., Citation2021). This helps farmers take targeted control measures, reduce the overuse of pesticides, and lower production costs (Badrinarayanan et al., Citation2017; Mattihalli et al., Citation2021).
Deep neural networks possess formidable learning and training capabilities, comprehensively acquiring image information of plant leaf diseases. This introduces a novel technique for identifying and detecting pear tree leaf diseases, enabling timely implementation of corresponding preventive measures and effectively mitigating the impact of conditions on crop yield and quality. Wang et al. (Wang et al., Citation2023) proposed the MFBP-UNet model, a novel approach based on the UNet architecture. It comprises a multiscale feature extraction (MFE) module and a tokenized multilayer perceptron (BATok-MLP) module with dynamic sparse attention. Experimental results demonstrate that, compared to other segmentation networks, MFBP-UNet exhibits outstanding performance, achieving significant improvements across multiple metrics. Zhenzhen Cheng et al. (Citation2023) introduced an enhanced LA-dpv3+ method, integrating an attention mechanism module into the encoder phase of the DeepLabV3+ architecture with a MobileNetv2 backbone. The model achieves an accuracy of 89%. Zhao et al. (Citation2018) proposed a practical deep learning framework, utilizing an enhanced CNN for pixel-level mask region automatic detection and segmentation of plant diseases. Yuan et al. (Citation2022) introduced an improved DEEPLabv3+ deep learning network, incorporating ResNet101 as the Backbone and a channel attention module in the remaining modules. Additionally, a feature fusion branch based on the feature pyramid network was introduced in the encoder to integrate features from different levels. Through these enhancements, the network’s evaluation metrics showed significant improvement, particularly achieving substantial progress in enhancing the segmentation of grape black rot lesions. Ru Jiaqi et al. (Citation2023) employed UNet++ for segmenting grape black spot lesions, achieving a segmentation accuracy of 97.41%. Zhong et al. (Citation2020) proposed an algorithm based on Mask-RCNN for segmenting and recognizing multiple target leaves in complex backgrounds, with a segmentation accuracy of 97.51%. Mao et al. (Citation2023) introduced an improved UNet model incorporating attention mechanisms and a CNN-Transformer hybrid structure. In handling complex backgrounds, pixel segmentation accuracy reached 92.56%. Chen et al. (Citation2017) presented the DeepLab series of networks, where DeepLabV3 introduced a significant number of dilated convolutions in the encoding stage, expanding the receptive field to include more information in each convolutional output. Dai Yushu et al. (Citation2021) utilized dilated convolution technology to establish a wheat scab detection and recognition model based on the DeepLabV3+ network, validated and evaluated with measured data, achieving an average intersection-over-union (mIoU) of 79%. Huang et al. (Citation2021) proposed using a semantic segmentation model based on the phase correlation algorithm and U-Net for foreground segmentation in field lettuce multispectral images. The canny algorithm was employed for edge extraction in multispectral channel images, resulting in an average segmentation accuracy of 94.98%. Although the Deeplabv3+ model is widely applied in satellite imagery, medical imaging, urban landscape segmentation, and element extraction (Das et al., Citation2021; Hu et al., Citation2022; Ono et al., Citation2021; Peng et al., Citation2020; Yu et al., Citation2022), its application in plant diseases is relatively limited. They adopted the lightweight MobileNetV2 as the Backbone to reduce the computational load, making the model suitable for embedding in mobile devices. The introduction of attention mechanisms aims to enhance the performance of neural networks in semantic segmentation tasks. Inspired by the advantages of Deeplabv3+ (Xue et al., Citation2023), we constructed an improved Deeplabv3+ for image segmentation of pear tree leaf diseases.
While various advanced models have emerged in plant disease identification, achieving significant accomplishments, substantial challenges still exist. In the segmentation task, the existing models also show diversity in segmentation performance for different data sets, tiny lesion characteristics, blurred boundaries, and changing shapes. An improved method for pear tree leaf disease segmentation based on DeepLabv3+ is proposed to address these challenges. The primary contributions are as follows:
Collect and prepare images of pear leaf diseases, partitioning the data into training and validation sets. The detailed data collection process is outlined in section 1.1.
Introduce the SE module to modify the pyramid pooling section of DeepLabv3+. This reduction in the model’s computational complexity contributes to the refined segmentation of pear tree leaf diseases. Experimental findings demonstrate a 4.3% enhancement compared to the baseline DeepLabv3+.
Substitute the original Xception model in DeepLabv3+ with the MobileNetv2 module, significantly accelerating the detection speed of the network model.
2. Method
2.1. Dataset construction
In this project, a dataset of pear leaf diseases was independently constructed. Image collection took place in the pear orchard at Tarim University, situated in the western region with a climate characterized by typical high-temperature seasons conducive to the healthy growth of pear tree leaves. Due to various factors influencing image capture, such as natural light and the photography device, data was collected between June 2023 and October 2023, utilizing a Huawei P40 device with a resolution of 3456 × 4608 pixels. The collected dataset includes photos taken in overcast and well-lit conditions to represent the environmental conditions authentically.
2.2. Example dataset
The study primarily followed the principles of multi-class, uniform distribution, and precise imaging to select disease images. The goal was to ensure that the research dataset encompasses a diverse range of samples, including large lesions, connected lesions, densely clustered small lesions, and faint lesions, each presenting varying degrees of difficulty for segmentation. The dataset included three types of pear tree leaf diseases: Brown Spot Disease, roll gall stripe Disease, and Black Spot Disease, along with images of healthy leaves, the details are shown in .
2.2.1. Dataset expansion
We performed data augmentation to address the challenge of a relatively limited dataset and the imbalance in label distribution for pear tree leaf diseases, which may lead to overfitting during training. This further enhanced the model’s generalization, sample diversity, and robustness. Through data augmentation, we could emphasize the features of disease spots, thereby improving the model’s learning capacity and accuracy.
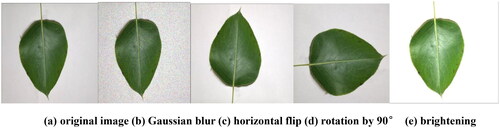
The data augmentation operations included Gaussian blur, which is beneficial in real-world applications where noise might affect data, such as changes in lighting conditions and errors in imaging devices. Data augmentation helps the model adapt better to such noise. Brightness adjustment, involving the modification of image brightness, simulates various lighting conditions. Image rotation, achieved by rotating images, allows the model to better adapt to objects at different angles. Flipping, either horizontally or vertically, enhances the model’s adaptability to image orientation. The effects of some of these operations are illustrated in .
Figure 2. Examples of data augmentation. (a) original image; (b) Gaussian blur; (c) horizontal flip; (d) rotation by 90°; (e) brightening.
Therefore, by augmenting the original images of pear tree leaf diseases, including 232 images of black spot disease, 116 images of brown spot disease, 88 images of pear vein yellows disease, and 65 images of healthy leaves, we aimed to diversify the dataset. The augmentation details are presented in . The augmented dataset comprised a total of 2004 images of pear tree leaf diseases, divided into training (1602), validation (201), and test (201) set in an 8:1:1 ratio.
Table 1. Dataset.
2.2.2. Dataset annotation
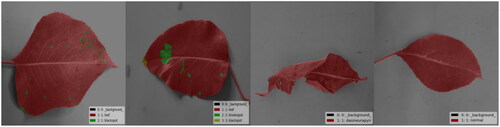
Processing the images involves adjusting them to a resolution of 512 × 512, performing detailed annotation, and Analyzing the disease spots on pear tree leaves. A labeling tool is utilized to precisely mark the areas of disease spots on the pear tree leaves, storing this annotation information in JSON format. Subsequently, the labeled data is transformed into a PNG image format for further processing. presents examples of partially annotated images. Six main categories were used for labeling: Label 1 represents the background; Label 2 represents Pear Tree Leaf Black Spot Disease; Label 3 represents Brown Spot Disease; Label 4 represents roll gall stripe Disease; Label 5 represents Healthy Leaves; and Label 6 represents Leaf Contours. Annotate the contours on the same leaf and different lesions to enable precise segmentation in experiments, allowing identification and classification.
2.3. Pear tree segmentation model
2.3.1. DeepLabv3+ model
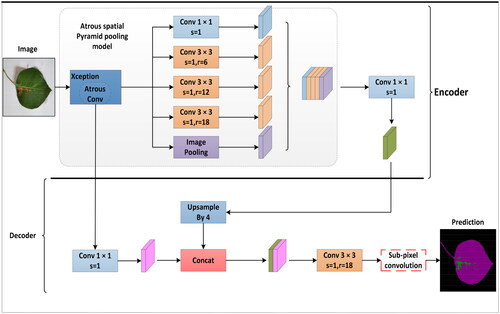
DeepLabv3+ has demonstrated outstanding performance in various fields with its robust, fully convolutional neural network (FCN) encoder-decoder structure. The model has been successfully applied to segmenting roads, vehicles, and pedestrians in autonomous driving, providing reliable visual understanding for intelligent transportation systems (Azad et al., Citation2020). Similarly, DeepLabv3+ has achieved significant organ structure segmentation and localization results in medical imaging, offering precise image analysis support for medical image diagnosis. In map-making, the model can extract essential information such as urban structures, green areas, and roads from satellite images, providing efficient data processing tools for urban planning and geographic information systems (Baheti et al., Citation2020). For segmenting disease spots on pear tree leaves, the encoder of DeepLabv3+ plays a crucial role in extracting features related to the morphological distribution of plant diseases. This encoder can leverage the ASPP module to capture contextual information at different scales, enabling a more comprehensive treatment of pear tree leaf diseases of various sizes and shapes and enhancing the model’s understanding of image semantics. Through upsampling operations, the decoder restores the feature maps extracted by the encoder to the resolution of the input image, ultimately generating accurate semantic segmentation results. The network architecture is depicted in . This study thoroughly analyzes the superior performance of DeepLabv3+ in specific tasks, providing robust methods and theoretical support for the Analysis of plant disease images.
2.3.2. Mobilenetv2 backbone network
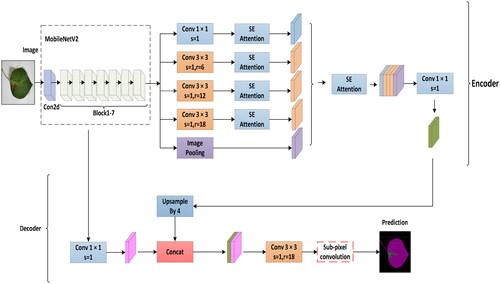
In deep learning, the DeepLabv3+ model has demonstrated outstanding precision in semantic segmentation tasks. However, there are problems with the Xception backbone network used in the traditional DeepLabv3+ model, which will lead to complexity and performance degradation. This study, focusing on the semantic segmentation of pear tree leaves, addresses these concerns by introducing the lightweight MobileNetv2 as a replacement for Xception. The structural parameters of MobileNetv2 are detailed in . This improvement aims to enhance computational efficiency, reduce complexity, and achieve superior performance in pear tree leaf disease segmentation tasks (Sandler et al., Citation2018). MobileNetv2, proposed by Google, incorporates post-activation after convolution, utilizing a linear bottleneck structure, residual connections, and an inverted residual structure to enhance the network’s non-linear representation capability (Dong et al., Citation2020). Improvements to the residual design involve adjusting the order of dimensionality reduction and increase, employing a 3 × 3 depthwise convolution, making it adaptable to varying resource constraints, and allowing users to adjust the model size dynamically.
Table 2. Mobilenetv2 structure parameter table.
Regarding feature extraction, the neural network extracts relevant information from the target and embeds it into a low-dimensional space. The reactivation function in the linear bottleneck structure is transformed into a linear function to reduce the loss of helpful network information. These enhancements and adjustments improve the accuracy and adaptability of pear tree leaf semantic segmentation, making the model more practical.
2.3.3. SE attention mechanism
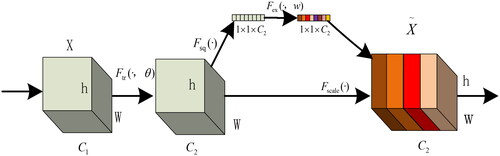
The fundamental concept of the SE module involves applying the Squeeze operation to the feature map post-convolution, aiding in obtaining comprehensive features at a global channel level (Hu et al., Citation2018). The model captures inter-channel correlations through the Excitation operation, obtaining weights for each channel. Subsequently, these weights are applied to the original feature map of pear tree leaf disease spots, resulting in the final feature map while preserving the input feature map size of the pear tree leaf image. In practical applications, the SE module, through channel-wise execution of attention or gating operations, utilizes a fully connected layer to expand the features originally mapped in low-dimensional space to a higher-dimensional space, recalculating the weight of each feature map. This process enables the model to understand and express the relationships between different channels more precisely, enhancing the effective representation of input data features. Ultimately, these weights are amalgamated to form a distribution that signifies the importance of elements in the feature map. The structure of the SE attention mechanism is illustrated in , where different colors represent distinct numerical values used to measure channel importance (Jin et al., Citation2022). The diversity of pear tree leaf lesions may affect segmentation results when extracting features in traditional methods. The SE attention mechanism adaptively selects and enhances critical feature channels within pear tree leaf lesions while suppressing unimportant media. This improves the network’s representation of input data features, strengthening its distinctiveness.
2.3.4. Optimizing the loss function
DeepLabv3+ is commonly employed for pixel-level segmentation tasks, typically utilizing cross-entropy loss as the primary loss function. A linear combination of cross-entropy loss (CELoss) and Dice loss is generally employed to enhance performance. In pixel-level segmentation tasks, background pixels often far outnumber defective samples. Solely relying on cross-entropy loss may lead to the network getting trapped in the issue of local minima (Botev et al., Citation2013). To address this concern, introducing the Dice loss function measures sample similarity, particularly emphasizing the correct classification of foreground images without overly stressing background pixels (Keskar & Socher, Citation2017). The approach effectively addresses the challenge of imbalanced foreground and background samples. By synergistically applying Dice loss and cross-entropy loss and employing a strategy with a non-linear loss function, it robustly combats the extreme imbalances in the data. This strategy prioritizes variations in the loss of the target region, aiming to mitigate the impact of feature region size on segmentation accuracy. By emphasizing the overall image loss and accurate loss calculations in the target region, the objective is to enhance segmentation precision. This approach takes into account a global perspective in achieving improved accuracy.
The loss rate Lce of the CELoss function is:
(1)
(1)
The loss rate LDice of the Dice loss function is denoted by:
(2)
(2)
In this equation, |X∩Y| represents the intersection of sets X and Y, while |X| and |Y| denote the number of pixels in locations X and Y, respectively. N represents the total number of pixels in the segmented image, and n represents the number of training iterations each time. Additionally, xn represents the independent variable for loss function training, while yn represents the dependent variable for gradient changes.
2.3.5. Improved lightweight DeepLabv3+ network model
In the conventional DeepLabv3+ network architecture, an improved Xception perception network is chosen for feature extraction. ASPP achieves feature extraction at different spatial scales by introducing dilated convolution operations. Due to the large number of parameters and convolution operations in the network layers, the model’s complexity increases, leading to challenges in training and slow convergence. To enhance network segmentation performance and movement efficiency, three improvements were made to the pear tree leaf disease image segmentation network based on the improved DeepLabv3+: (1) The conventional DeepLabv3+ employs the Xception perception network, but its extensive parameters and convolutional operations pose training difficulty and slow speed challenges. To address this, we propose replacing Xception with MobileNetV2 to reduce model complexity, minimize the number of parameters, and alleviate the risk of overfitting. Simultaneously, we aim to maintain effective feature extraction while reducing the network’s overall size. The lightweight characteristics of MobileNetV2 enable efficient training within a relatively short time, facilitating better generalization to unseen data. This enhances the model’s ability to generalize well on input data, making it more suitable for efficient image segmentation in embedded devices and mobile applications. (2) Introduce the SE module after the ASPP module in MobileNetV2 to emphasize channel weight learning, adaptively focusing on essential features and suppressing irrelevant information. The SE module is incorporated after each ASPP convolution to comprehensively capture contextual information, enhancing the segmentation model’s perceptual capabilities. This improvement exhibits significant advantages in boosting segmentation performance and global awareness while mitigating the impact of overfitting. It aids the model in better learning disease features, thereby improving its generalization ability. (3) Optimize the loss function by introducing cross-entropy loss to address sample imbalance issues. Adopting a linear combination of cross-entropy and Dice loss enables a more careful consideration of overall image loss and accurate loss calculation for target regions, further enhancing segmentation accuracy. The specific improvements are illustrated in . These enhancements effectively address challenges that conventional loss functions may encounter in handling sample imbalance, aiming to improve performance, achieve real-time capabilities, and effectively address sample imbalance issues, providing robust support for more accurate segmentation of pear tree leaf diseases in practical applications.
3. Model training and analysis of experimental results
3.1. Experimental environment configuration
During the experimental process, to ensure uniformity, model training was conducted in the same environment, with specific configurations outlined in the .
Table 3. Table of experimental environment parameters.
3.2. Training setup
During the experimental process, to ensure the comparability of network performance, comparative experiments were conducted using the same dataset under identical environmental conditions. Experimental parameters were configured, maintaining a consistent image size set at 512 × 512 pixels for uniform processing. To avoid insufficient GPU memory due to a large batch size, which could lead to unstable network training, we constrained the batch size to 4 and conducted a total of 300 iterations. This setting was determined through experimental results, aiming to strike the optimal balance between efficiency and accuracy. As different learning rates may result in variations in loss values, a lower learning rate can slow down convergence. Hence, we set the learning rate to 5e-4. To meet the model’s requirements at different training stages, we employed the Adam (Tiwari et al., Citation2020) optimizer, adjusting network parameters to minimize the loss function and expedite model convergence. These experimental configurations facilitate the training and optimization of deep learning models. For specific parameter settings, refer to the .
Table 4. Experimental parameter settings.
3.3. Model evaluation metrics
PA is the ratio of correctly predicted pixels to the total number of a particular class. PA represents the average of PA for each category. Where Pi is the pear leaf disease display prediction sample, and PiPij is the pear leaf disease error prediction sample. Where N is the pear tree leaf disease type, TP is the correct prediction sample of pear tree leaf disease, FP is the wrong prediction sample, and FN is the undetected pear tree leaf disease prediction sample.
The mathematical expressions of PA and MPA are as follows:
(3)
(3)
(4)
(4)
(5)
(5)
(6)
(6)
(7)
(7)
(8)
(8)
3.4. Results and analysis
Utilizing the refined Deeplabv3+ network model, 300 iterations of training were conducted on the pear tree leaf disease dataset to acquire a thoroughly trained and effective model. Through this training process, not only was the model’s training speed accelerated, but there was also a significant enhancement in the model’s capacity to fit the data, thereby improving the accuracy of lesion segmentation on the limited pear tree leaf disease dataset. Through this series of steps, a well-trained and optimized model was successfully constructed, demonstrating efficiency in accomplishing the task of pear tree leaf disease segmentation in practical applications.
3.4.1. Comparison of the traditional model and improved model
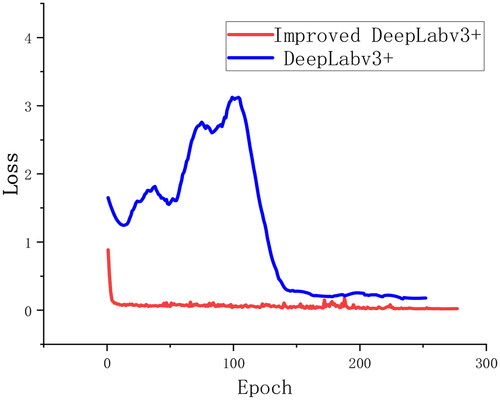
To validate the model’s performance, segmentation experiments were conducted on the pear tree disease training set. presents the training results of the Deeplabv3+ model and its improved version on the validation set, where the blue line represents the training process of the traditional Deeplabv3+ model. In contrast, the red line signifies the training progress of the enhanced Deeplabv3+ model. Observing the graph, it is evident that after 150 iterations, the loss values of Deeplabv3+ gradually stabilize. With an increase in the number of iterations, the model’s loss values progressively decrease and stabilize. Compared to the traditional Deeplabv3+ model, the improved version exhibits a faster convergence rate and more minor fluctuations, stabilizing around 25 iterations. This is primarily attributed to introducing the SE attention mechanism, effectively incorporating channel-wise dependencies, integrating global contextual relationships, capturing contextual feature information, significantly enhancing feature representation, and thereby improving the effectiveness of network learning.
3.4.2. Verification of different backbone network
The core objective of the experiment is to delve into and assess the effectiveness of replacing Xception with MobileNetV2 as the backbone network for Deeplabv3+ in the semantic segmentation task of pear tree leaf disease images. Due to the outstanding performance of lightweight backbone networks in resource-constrained environments, this substitution is potentially considered to enhance the efficiency of the segmentation model. The experimental design, Xception, and MobileNetV2 are compared under the same conditions. Collecting and comparing key metrics such as accuracy, Recall, MIoU, etc., makes it possible to evaluate this replacement’s impact on semantic segmentation performance objectively. This comparison contributes to understanding the applicability of lightweight backbone networks in specific tasks and provides a foundation for selecting an appropriate network architecture. The particular metrics are outlined in .
Table 5. Comparison of influence of different backbone networks.
The experiment demonstrates the superiority of different backbone networks within the Deeplabv3+ framework in the segmentation task of pear tree leaf diseases. The results in indicate that altering the backbone network significantly influences model performance, with all performance metrics showing relative improvement. The replacement of Xception with MobileNetV2 brings dual advantages to the network backbone. Firstly, this substitution greatly enhances the algorithm’s real-time capabilities, making it more suitable for real-time or edge device applications. Secondly, due to the lightweight nature of MobileNetV2, the model’s volume is significantly reduced while maintaining performance, improving its efficiency. This is particularly valuable in resource-constrained scenarios, such as mobile devices or edge computing platforms. Regarding segmentation speed, the experimental results show an approximately 5-fold increase compared to Xception after replacing the backbone network and it’s also faster than Xception, and it’s also faster than Xception. This confirms the significant advantage of MobileNetV2 in improving segmentation speed, providing robust support for efficient segmentation of pear tree leaf diseases in resource-constrained environments.
3.4.3. Validation of different optimizers
Two tests were primarily conducted using SGD and Adam optimizers to validate the rationality of choosing different optimizers. As shown in , in the original model, the use of the SGD optimizer did not lead to an improvement in MIoU; instead, it caused a decrease to 78.31%. On the other hand, employing the Adam optimizer increased data performance. Similarly, when replacing the backbone network, using the SGD optimizer did not enhance the data; instead, it decreased MIoU to 79.32%. Experimental results demonstrate that, compared to the memory-intensive SGD optimization algorithm, Adam exhibits smaller memory usage. The stochastic nature of the SGD optimizer allows it to quickly search for relatively good features in large-scale data, making it suitable for scenarios with limited resources. However, SGD’s convergence speed is relatively slow, leading to a decrease in performance. Adam, on the other hand, demonstrates stable performance without the need for manual adjustment of the learning rate across different network structures and datasets. Adam usually provides more stable performance, with stronger adaptability in terms of adaptive learning rates and better robustness to hyperparameters. This reduces sensitivity to hyperparameters, making it easier to use the pear tree disease model in practical applications and, to some extent, improving model stability. Additionally, given the small size of the pear tree disease dataset, Adam’s introduction of adaptive learning rates and momentum contributes to superior performance across different scenarios of pear tree leaf disease.
Table 6. Comparison of the effects of different optimizers.
3.4.4. Validation of different attention mechanisms
The choice for the main backbone network is MobileNetV2, considering its lightweight structure and versatility. The SE attention mechanism effectively suppresses irrelevant information features. In the segmentation of small targets such as leaf disease, the MobileNetV2 model has limitations in detecting features of small targets. Therefore, the addition of attention mechanisms addresses this limitation. Specifically, four different attention modules were studied, including SE, Efficient Channel Attention (ECA) (Tiwari et al., Citation2020; Wang et al., Citation2020), Convolutional Block Attention ModuleCBAM (CBAM) (Du et al., Citation2021; Woo et al., Citation2018), and Channel Attention (CA) (Fang et al., Citation2022; Zhang et al., Citation2018). presents the performance of various modules on the validation set across different metrics. This comparative Analysis helps identify which module introduction is more crucial for performance improvement in a specific task. This, in turn, provides more targeted recommendations for the model design of pear tree leaf disease segmentation tasks, offering a more comprehensive evaluation of the proposed improvement scheme and enhancing its practical applicability. The details are shown in .
Table 7. Comparison and verification of different attention mechanisms.
From , it can be observed that by replacing the backbone network of DeepLabv3+ for pear tree leaf disease segmentation, the model achieved impressive segmentation performance with MIoU, Recall, Precision, and F1-score reaching 86.32%, 88.97%, 91.10%, and 88.97%, respectively. Looking at MIoU specifically, the introduction of CA, ECA, CBAM, and SE attention mechanisms resulted in 82.32%, 83.79%, 84.02%, and 86.32%, respectively, showing a 2.55% improvement compared to not incorporating attention mechanisms. When considering various metrics comprehensively, the SE attention mechanism demonstrated outstanding performance in MIoU and achieved good overall performance in other metrics. For instance, the average time per iteration was 61.80 seconds, saving 2.04 seconds per iteration compared to not using attention mechanisms. The experiment demonstrated that introducing attention mechanisms did not increase the computational load of the model.
Additionally, while ensuring segmentation accuracy, it reduced the computational load. This highlights the superiority of the SE attention mechanism in pear tree leaf disease segmentation tasks, providing a solid reference for the design of models in similar schemes. Future research could further compare the adaptability of different attention mechanisms across other datasets and tasks, advancing the development of attention mechanisms in deep learning segmentation.
3.4.5. Experimental comparison of different models
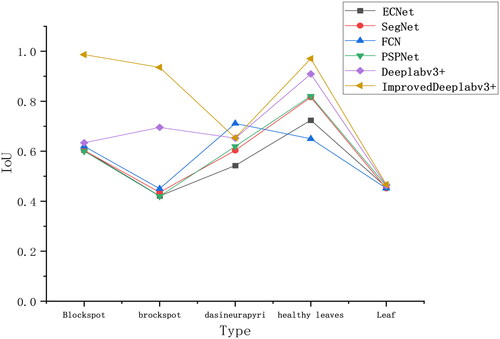
For a more intuitive assessment of the model’s Intersection over Union (IoU) for pear tree leaf disease spots, compares the segmentation IoU of five different network models (CENet, SegNet, FCN, PSPNet, and DeepLabv3+) with the proposed network. From the figure, it is evident that other models exhibit varying segmentation performance for different labels. For example, CENet demonstrates good segmentation performance for more significant features but performs poorly in segmenting small disease spots and irregular leaf contours. SegNet, FCN, and PSPNet models exhibit suboptimal segmentation performance for small disease spots and diverse, complex leaf contours. The suboptimal segmentation performance is mainly attributed to the small and dense nature of black spot disease spots and the annotation of leaf contours during labeling, leading to different disease spots on different leaf contours, resulting in poor feature learning by the model.
In contrast, the DeepLabv3+ model performs well in segmenting healthy leaves and prominently featured disease spots but shows average segmentation performance for the less represented curled leaf vein disease. As seen in , the improved DeepLabv3+ model demonstrates superior segmentation accuracy for pear tree leaf black spot disease, brown spot disease, roll gall stripe Disease, and healthy leaves. Its segmentation performance for small target black spot disease spots surpasses other models. In the pear tree leaf disease dataset, the improved accuracy of the proposed network model exceeds the other five networks, indicating its excellent segmentation capability for pear tree leaf disease datasets.
Through comparative experiments between the improved model and others, we further validate the efficacy and feasibility of the enhanced model. shows the test results of the leaf validation set.
Table 8. Comparison of segmentation effects of different network models.
As shown in , in terms of MIoU, the metrics for CENet, SegNet, FCN, PSPNet, and Deeplabv3+ are 41.43%, 78.27%, 81.14%, 81.51%, and 82.02%, respectively. The improved Deeplabv3+ outperforms all other models with an MIoU of 86.32%. The experiment indicates that CENet exhibits suboptimal segmentation performance on the pear tree leaf disease dataset, while SegNet, FCN, PSPNet, and Deeplabv3+ all fall short compared to the enhanced model. Overall, the improved Deeplabv3+ demonstrates outstanding segmentation accuracy based on the Analysis of these three metrics.
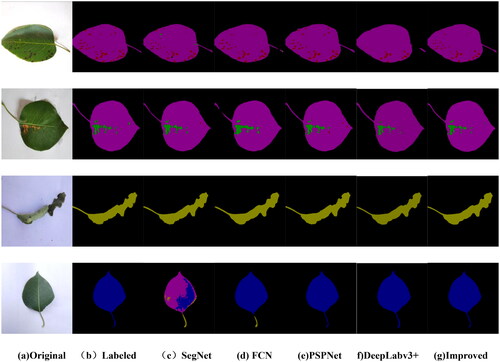
For a more intuitive comparison of the segmentation results of different networks, randomly selected images of pear leaf diseases, including brown spot disease, black spot disease, roll gall stripe Disease, and healthy pear leaves. The segmentation results on the test set of pear tree leaf images are presented in , showcasing outcomes achieved with different network architectures. The first column shows images of pear leaf diseases, the second column displays manually labeled disease images, and each subsequent column represents the segmentation results of different network models. Black indicates the background, pink represents the leaves themselves, red denotes segmented brown spot diseases, green represents segmented black spot diseases, brown signifies pear vein yellows, and blue represents healthy leaf areas. From , it is evident that different conditions exhibit distinct colors and shapes. Concerning the lesions on pear tree leaves, the segmentation results of SegNet reveal areas for improvement, notably instances of missegmentation and omissions. This is primarily attributed to the relatively small dataset of healthy leaves, resulting in an imbalance among the four label categories. The FCN network model can roughly segment the disease areas, but inaccuracies and missed segmentations are observed in some regions, especially for small black spot and brown spot diseases. PSPNet and DeepLabv3+ significantly improve segmentation results, aligning more closely with the labels for pear leaf diseases. However, PSPNet exhibits errors in segmented edges and other disease areas, while DeepLabv3+ has less clear boundaries. For the improved DeepLabv3+ network, the segmented disease areas closely match the labeled regions, even capturing tiny lesions on the diseased leaves. This suggests that the improved DeepLabv3+ accurately segments disease images, including the disease regions and their boundaries. This advantage arises from utilizing MobileNetV2 as the backbone network and incorporating the SE attention mechanism in the ASPP module. This design accentuates the feature information of pear tree leaf disease spots while simultaneously suppressing features irrelevant to the task. Such a design has resulted in higher efficiency during the re-segmentation stage. In summary, compared to other networks, the improved DeepLabv3+ demonstrates superior performance in the segmentation task of pear tree leaf diseases, highlighting the suitability of its adopted network model for this specific application scenario.
4. Discussion
This study proposes an improved DeepLabv3+ model for segmenting pear leaf diseases based on the DeepLabv3+ framework. Three types of conditions and healthy leaf segments are tested. The ASPP is a crucial encoder component in semantic segmentation tasks, utilizing multiple parallel dilated convolutions to extract multi-scale information. These features play a vital role in determining the segmentation target locations and boundaries, enhancing the model’s perceptual capabilities for various sizes and shapes and thereby improving segmentation performance. Replacing the Xception module with the MobileNetV2 module in the DeepLabv3+ model enables its practical application in embedded systems, reducing the computational burden and semantic segmentation workload. Cheng et al. (Citation2023) consistently achieved improved results by replacing the backbone with MobileNetV2, enhancing the computational speed of the model. In the experiments, the Xception module in the DeepLabv3+ model was substituted with the MobileNetV2 module, enabling its effective utilization in embedded systems. This modification significantly reduces the computational burden of the model and decreases the workload for lightweight semantic segmentation. As a result, the average computation is increased by nearly five times compared to the original backbone network. By introducing the SE attention mechanism, the model can focus on task-relevant feature information while suppressing irrelevant details. This design not only enhances the accuracy of semantic segmentation but also improves the stability of the model in different scenarios. Experimental results indicate that the improved DeepLabv3+ model achieves pixel accuracy, MIoU, recall, precision, and F1-score of 86.32%, 88.97%, 91.10%, 88.97%, respectively. In comparison, Cai et al. (Citation2022) reported an MIoU metric of 81.3%, demonstrating the superiority of our proposed model. In summary, the enhanced DeepLabv3+ model exhibits superior performance in the classification of pear leaf disease segmentation, making it a valuable contribution to practical applications in orchards.
While the current task showcases the network model’s primary strengths in outstanding performance and rapid operational speed, it is acknowledged that future research holds significant room for improvement. Specifically, in practical applications, the adoption of a comprehensive training strategy considering both image quality and environmental factors remains suboptimal. Utilizing data augmentation for model training, future considerations may involve the implementation of transfer learning or domain adaptation methods. This approach aims to enhance the model’s adaptability to different environments, thereby elevating its robustness. The diverse array of leaf types leads to suboptimal segmentation results. Further optimization of the network structure is necessary to accurately segment leaves of varying shapes. Alternatively, exploring superior semantic segmentation models (Tiwari et al., Citation2020; Yang et al., Citation2020; Bhagat et al., Citation2022; Divyanth et al., Citation2023) could enhance adaptation to complex natural environments. Limitations persist during training. While the replacement of the Xception module with MobileNetV2 reduces computational complexity, it may compromise feature learning capabilities. Introducing the SE attention mechanism after ASPP aims to augment the model’s feature learning, enhancing overall performance. Further refinement of the MobileNetV2 structure, without introducing attention mechanisms, should be considered to improve the extraction of complex features. Inspired by Zhang et al. (Citation2023) work, introducing a lightweight DeepLabV3+ network-based image processing pipeline for estimating various lettuce traits (fresh weight, dry weight, plant height, diameter, leaf area) in complex backgrounds has proven effective. Further optimizing the network model for specific scenarios, training on different plant diseases and types, is essential to enhance the model’s generalization capabilities.
5. Conclusion
In the enhanced network architecture, integrating MobileNetV2 aims to maintain feature extraction effectiveness while reducing the model’s parameter count, thereby improving segmentation speed. This refinement renders the model more suitable for real-time applications and resource-constrained scenarios like edge computing and embedded systems. Introducing the SE attention mechanism after the ASPP module enables the network to focus more on critical areas, thus improving the segmentation accuracy of pear tree leaf diseases. Incorporating the SE attention mechanism facilitates the adaptive learning of channel weights, concentrating on essential features while suppressing task-irrelevant information, thereby strengthening the perception of the diseased areas. Experimental results demonstrate that the improved model achieves MIoU, mPA, mPr, and mRecall scores of 86.32%, 88.97%, 91.10%, and 88.97%, respectively, marking a 4.3% improvement in MIoU compared to traditional DeepLabv3+.
Moreover, it outperforms segmentation methods such as CENet, SegNet, FCN, and PSPNet. This underscores the enhanced model’s superiority and hints at its potential applications in practical scenarios. Future research directions may involve exploring strategies to simplify the model further and considering the severity of lesions to address pear tree leaf disease issues comprehensively. Additionally, integrating the solution into the latest disease detection platforms can enhance the method’s feasibility and scalability for practical production applications. This objective aims to achieve higher segmentation accuracy and efficiency in real-world applications, providing a more precise tool for plant disease monitoring in agriculture.
Authors’ contributions
Writing—original draft preparation, J.F.; resources, X.L.; tidying up, F.C.; supervision G.W. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
The authors would like to show sincere thanks to those technicians who have contributed to this research.
Disclosure statement
The authors declare no conflicts of interest.
Data availability statement
The experimental data used to support the findings of this study are available from the authors upon request.
Reference
- Azad, R., Asadi-Aghbolaghi, M., Fathy, M., & Escalera, S. (2020). Attention deeplabv3+: Multi-level context attention mechanism for skin lesion segmentation. European conference on computer vision (pp. 1–16). Springer International Publishingvolctrans extension anchor for patchId=2727.
- Badrinarayanan, V., Kendall, A., & Cipolla, R. (2017). SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12), 2481–2495. https://doi.org/10.1109/TPAMI.2016.2644615
- Baheti, B., Innani, S., Gajre, S., & Talbar, S. (2020). Semantic scene segmentation in unstructured environment with modified DeepLabV3+. Pattern Recognition Letters, 138, 223–229. https://doi.org/10.1016/j.patrec.2020.07.029
- Bhagat, S., Kokare, M., Haswani, V., Hambarde, P., & Kamble, R. (2022). Eff-UNet++: A novel architecture for plant leaf segmentation and counting. Ecological Informatics, 68, 101583. https://doi.org/10.1016/j.ecoinf.2022.101583
- Botev, Z. I., Kroese, D. P., Rubinstein, R. Y., & L’ecuyer, P. (2013). The cross-entropy method for optimization in handbook of statistics. Elsevier, 31, 35–59.
- Cai, M., Yi, X., Wang, G., Mo, L., Wu, P., Mwanza, C., & Kapula, K. E. (2022). Image segmentation method for sweetgum leaf spots based on an improved DeeplabV3+ network. Forests, 13(12), 2095. https://doi.org/10.3390/f13122095
- Chen, L. C., Papandreou, G., Schroff, F., et al. (2017). Rethinking atrous convolution for semantic image segmentation. arXiv:1706.05587.
- Cheng, Z., Cheng, Y., Li, M., Dong, X., Gong, S., & Min, X. (2023). Detection of cherry tree crown based on improved LA-dpv3+ algorithm. Forests, 14(12), 2404. https://doi.org/10.3390/f14122404
- Das, S., Fime, A. A., Siddique, N., & Hashem, M. M. A. (2021). Estimation of road boundary for intelligent vehicles based on DeepLabV3+ architecture. IEEE Access, 9, 121060–121075. https://doi.org/10.1109/ACCESS.2021.3107353
- Divyanth, L. G., Ahmad, A., & Saraswat, D. (2023). A two-stage deep-learning based segmentation model for crop disease quantification based on corn field imagery. Smart Agricultural Technology, 3, 100108. https://doi.org/10.1016/j.atech.2022.100108
- Dong, K., Zhou, C., Ruan, Y., et al. (2020). MobileNetV2 model for image classification [Paper presentation]. 2020 2nd International Conference on Information Technology and Computer Application (ITCA) (pp. 476–480). IEEE. https://doi.org/10.1109/ITCA52113.2020.00106
- Donghui, S., Xiuliang, J. U., Dengchao, F., et al. (2016). Face recognition from aggregated images based on FAST detector and SURF descriptor. Foreign Electronic Measurement Technology, 35(1), 94–98.
- Du, W., Rao, N., Dong, C., Wang, Y., Hu, D., Zhu, L., Zeng, B., & Gan, T. (2021). Automatic classification of esophageal disease in gastroscopic images using an efficient channel attention deep dense convolutional neural network. Biomedical Optics Express, 12(6), 3066–3081. https://doi.org/10.1364/BOE.420935
- Fang, Y., Huang, H., Yang, W., Xu, X., Jiang, W., & Lai, X. (2022). Nonlocal convolutional block attention module VNet for gliomas automatic segmentation. International Journal of Imaging Systems and Technology, 32(2), 528–543. https://doi.org/10.1002/ima.22639
- Huang, L. S., ShaO, S., Lu, X. J., et al. (2021). Segmentation and registration of lettuce multispectral image based on convolutional Neural Network. Transactions of the Chinese Society for Agricultural Machinery, 52(9), 186–194.
- Hu, J., Shen, L., & Sun, G. (2018). Squeeze-and-excitation networks. Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 7132–7141).
- Hu, Z., Zhao, J., Luo, Y., & Ou, J. (2022). Semantic SLAM based on improved DeepLabv3+ in dynamic scenarios. IEEE Access, 10, 21160–21168. https://doi.org/10.1109/ACCESS.2022.3154086
- Jiaqi, R. U., Bin, W., Xiang, W., et al. (2023). Segmentation and classification of grape black rot lesion degree based on improved UNet++ model [J/OL]. Journal of Zhejiang Agricultural, 35(11), 2720–2730volctrans extension anchor for patchId=3423.
- Ji-Feng, L. (2022). Chest CT image segmentation method based on improved Snake model. Automation & Instrumentation, 32(11), 2059–2066.
- Jin, X., Xie, Y., Wei, X.-S., Zhao, B.-R., Chen, Z.-M., & Tan, X. (2022). Delving deep into spatial pooling for squeeze-and-excitation networks. Pattern Recognition, 121, 108159. https://doi.org/10.1016/j.patcog.2021.108159
- Keskar, N. S., & Socher, R. (2017). Improving generalization performance by switching from adam to sgd. arXiv preprint arXiv:1712.07628.
- Lincai, H., Zhiwen, W. A. N. G., Xiaobiao, F., et al. (2022). Edge detection of gesture image: Canny algorithm based on multi-direction and optimal threshold. Journal of Guangxi University of Science and Technology, 33(1), 71–77.
- Mao, W. J., Ruan, J. Q., & Liu, S. (2023). Strawberry disease semantic segmentation based on improved UNet with attention mechanism. Computer Systems & Applications, 32(06), 251–259.
- Mattihalli, C., Gared, F., & Getnet, L. (2021). Automatic plant leaf disease detection and auto-medicine using IoT technology. In Krause P., & Xhafa F. IoT-based intelligent modelling for environmental and ecological engineering (pp. 257–273). Springer.
- Ono, H., Murakami, S., Kamiya, T., & Aoki, T., 12–15 October (2021). Automatic segmentation of finger bone regions from CR images using improved DeepLabv3+ [Paper presentation]. Proceedings of the 2021 21st International Conference on Control, Automation and Automation and Systems (ICCAS) (pp. 1788–1791), Jeju, Korea.
- Peng, H., Xue, C., Shao, Y., Chen, K., Xiong, J., Xie, Z., & Zhang, L. (2020). Semantic segmentation of litchi branches using DeepLabV3+ model. IEEE Access, 8, 164546–164555. https://doi.org/10.1109/ACCESS.2020.3021739
- Sandler, M., Howard, A., Zhu, M., et al. (2018). Mobilenetv2: Inverted residuals and linear bottlenecks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 4510–4520).
- Semma, A., Hannad, Y., Siddiqi, I., Djeddi, C., & El Youssfi El Kettani, M. (2021). Writer identification using deep learning with FAST keypoints and Harris corner detector. Expert Systems with Applications, 184, 115473. https://doi.org/10.1016/j.eswa.2021.115473
- Tiwari, D., Ashish, M., Gangwar, N., et al. 4th International Potato leaf Diseases Detection using Deep Learning. (2020).
- Tiwari, D., Ashish, M., Gangwar, N., et al. (2020). Potato leaf diseases detection using deep learning [Paper presentation]. 2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS) (pp. 461–466). IEEE. https://doi.org/10.1109/ICICCS48265.2020.9121067
- Wang, J. P. (2020). Main pear diseases and their control methods. Modern Horticulture, 43(07), 194–195.
- Wang, H., Ding, J., He, S., Feng, C., Zhang, C., Fan, G., Wu, Y., & Zhang, Y. (2023). MFBP-UNet: A network for pear leaf disease segmentation in natural agricultural environments. Plants, 12(18), 3209. https://doi.org/10.3390/plants12183209
- Wang, Q., Wu, B., Zhu, P., et al. (2020). ECA-Net: Efficient channel attention for deep convolutional neural networks. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 11534–11542).
- Wenbo, L., Tao, Y., & Qi, L. (2021). Tomato leaf disease detection method based on improved SOLO v2. Transactions of the Chinese Society for Agricultural Machinery, 52(8), 213–220.
- Woo, S., Park, J., Lee, J. Y., et al. (2018). Convolutional block attention module. Proceedings of the European Conference on Computer Vision (ECCV) (pp. 3–19).
- Xue, X., Luo, Q., Bu, M., Li, Z., Lyu, S., & Song, S. (2023). Citrus tree canopy segmentation of orchard spraying robot based on RGB-D image and the improved DeepLabv3+. Agronomy, 13(8), 2059. https://doi.org/10.3390/agronomy13082059
- Yang, K., Zhong, W., & Li, F. (2020). Leaf segmentation and classification with a complicated background using deep learning. Agronomy, 10(11), 1721. https://doi.org/10.3390/agronomy10111721
- Yanjun, H., Ting, Z., & Wu, X. J. (2021). Occurrence and prevention of several common pear diseases. Modern Rural Science and Technology, 52(9), 186–194.
- Yu, L., Zeng, Z., Liu, A., Xie, X., Wang, H., Xu, F., & Hong, W. (2022). A lightweight complex-valued DeepLabv3+ for semantic segmentation of PolSAR image. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 15, 930–943. https://doi.org/10.1109/JSTARS.2021.3140101
- Yuan, H., Zhu, J., Wang, Q., Cheng, M., & Cai, Z. (2022). An improved DeepLab v3+ deep learning network applied to the segmentation of grape leaf black rot spots. Frontiers in Plant Science, 13, 795410. https://doi.org/10.3389/fpls.2022.795410
- Yushu, D., Xiao-Chun, Z., Chengming, S., Jun, Y., Tao, L., & Liu, S. P. (2021). Identification of wheat scab based on image processing and Deeplabv3+ model. Chinese Journal of Agricultural Mechanization, 42(09), 209–215.
- Zhang, Y., Li, K., Li, K., et al. (2018). Image super-resolution using very deep residual channel attention networks. Proceedings of the European Conference on Computer Vision (ECCV) (pp. 286–301).
- Zhang, Y., Wu, M., Li, J., Yang, S., Zheng, L., Liu, X., & Wang, M. (2023). Automatic non-destructive multiple lettuce traits prediction based on DeepLabV3. Journal of Food Measurement and Characterization, 17(1), 636–652. https://doi.org/10.1007/s11694-022-01660-3
- Zhao, W., Zhang, H., Yan, Y., Fu, Y., & Wang, H. (2018). A semantic segmentation algorithm using FCN with combination of BSLIC. Applied Sciences, 8(4), 500. https://doi.org/10.3390/app8040500
- Zhong, W., Xinlei, L., Kunlong, Y., et al. (2020). Segmentation and recognition of multi-target blades under complex background based on Mask-RCNN. Acta Agriculturae Zhejiangensis, 32(11), 2059–2066.
- Zhongyuan, W., Ruimin, H., & Kai, Z. (2004). An implementation of region partition or connected component label split-and-merge method. Small Microcomputer System, 25(9), 1648–1651.