?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Kopyor coconut (Cocos nucifera L. family Arecaceae) is a mutant nut from Indonesia and Jember Regency serves as a center for the growth in the country. The population and morphology in this area have not been explored extensively to describe diversity. Therefore, this research aimed to determine the morphology and kinship of kopyor coconut in three sub-districts, namely Wuluhan, Ambulu and Gumuk Mas. A random sampling was conducted to select 10 trees from each dwarf (yellow and green dwarf kopyor (GKK and GHK)) and tall kopyor coconut (green, yellow and brown tall kopyor (DHK, DKK and DCK)). GPS-marked population was processed using GIS program to create a map for the distribution of kopyor coconut. Qualitative and quantitative data were collected on stems, leaves, flowers, fruits and endosperm. The results showed that there were 31 areas of kopyor coconut populations in Jember Regency. The high diversity was observed in the number of female flowers, as well as the percentage of kopyor fruit per bunch. DCK had the smallest and largest flower and fruit index values, respectively. In addition, the level of endosperm structure was classified into four types. Tall kopyor coconut had a low phylogenetic relationship of 29%, while DKK and DCK types reported the closest. This information was valuable in complementing the lack of data on the phenotype as well as the diversity of the population used for conservation, breeding programs, and production purposes.
Reviewing Editor:
1. Introduction
Kopyor coconut (Cocos nucifera L. family Arecaceae) is a genetically diverse coconut germplasm. Additionally, the palm has a natural genetic mutation passed down from parents to offspring in Indonesia (Sukendah & Volkaert, Citation2009). This mutation is expressed in endosperm, which differs from normal coconut. The majority of endosperm is not attached to the shell and breaks into small pieces mixed with coconut water. Therefore, this type is known as ‘kopyor’, meaning breaking into small pieces. Maskromo et al. (Citation2013) described kopyor endosperm as soft, fluffy and crumbly, which was released from the shell due to a lack of the enzyme a-D-galactosidase (Sukendah & Volkaert, Citation2009). Previous research reported that the trait was controlled by a recessive gene (Novarianto et al., Citation2014; Setiawan et al., Citation2020).
Mutant coconuts that resemble kopyor are also found in several countries. In the Philippines, Macapuno type has been reported with a soft, viscous, white, and translucent jelly-like endosperm (Sivakumar et al., Citation2022). Even though several investigations have argued that kopyor coconut is Indonesian Macapuno, the gelatinous characteristics of the endosperm are significantly different. According to Wicaksono et al. (Citation2021), the endosperm is very similar to a local mutant coconut in Banten, which is called ‘wax coconut’ (‘kelapa lilin’). Other similar mutants with local names are ‘Dikiri’, ‘Maprao Kathi’, ‘Thairu thengai’, ‘Sap’ from Sri Langka, Thailand, India and Vietnam, respectively (Gunathilake & Samarasinghe, Citation2015; Chomchalow, Citation1999; Jerard et al., Citation2013; Toan & Thanh, Citation2011).
Kopyor is like normal coconut, which is grouped into two major types, namely ‘Tall’ and ‘Dwarf’ based on morphological characteristics, growth and breeding behavior. Tall coconut shows mainly cross-pollination and late flowering ranging from 8 to 10 years after planting. Meanwhile, dwarf type is characterized by self-pollination and early flowering around 4–6 years after planting (Jerard et al., Citation2015; Xiao et al., Citation2017). Coconuts are also classified into three types namely tall, dwarf and hybrid or intermediate (var. aurantiaca), with the hybrid resulting from cross-pollination between tall and dwarf types (Swaminathan & Nambiar, Citation1961; Manimekalai & Nagarajan, Citation2010).
There are two classifications of kopyor coconut palms based on the type of fruit, namely heterozygous and homozygous. The heterozygous types bear both normal and kopyor coconut fruits in one bunch. These palms are commonly found in nature and are typically propagated from normal coconut seeds that bear kopyor gene. On the other hand, homozygous kopyor coconut produces 100% kopyor fruits in one bunch. This particular type has not been found in nature due to it is propagated through in vitro methods using embryo rescue techniques.
In Indonesia, kopyor coconut is used commercially for fresh drinks and ten times the price of the normal type on the market. However, the production mainly depends on coconut in central areas such as Pati and Sumenep in Central and East Java, respectively. Efforts to develop the population and production of kopyor coconut fruits are constrained by the lack of data on the existence, types and current availability.
Some investigations reported that the kopyor population is dispersed across several areas, including Sumenep, Jember, Pati, Ciomas and South Lampung (Sukendah et al., Citation2011; Maskromo et al., Citation2014). Sumenep is popularly known for tall type, showing morphologically distinct fruit types based on stems, leaves, flowers and fruit (Sukendah et al., Citation2011). Pati predominantly consisting of dwarf types are classified into three based on morphological characters, which are used as a standard for the release (Maskromo et al., Citation2016). However, there has been no exploration of kopyor coconut morphology in Jember Regency, increasing the difficulty of identifying the population, diversity, and phylogenetic relationship.
The population data, plant diversity and relationship among the types need to be investigated due to the existence of kopyor palms in Jember. According to Kurniasih et al. (Citation2020), plant diversity can be analyzed morphologically by direct observation of phenotypes or by using molecular markers. A phenotypic method is used to investigate phylogenetic relationships, which play a significant role in the efficient selection of parents in plant breeding programs for crosses (Amzeri et al., Citation2011). By identifying and selecting kopyor palms with superior characteristics, these individuals can be used as parents or mother trees.
This research aimed to examine the morphology of kopyor coconut from Jember, including the phenotypic diversity and phylogenetic relationship. The results provide early information data on the germplasm for a further conservation strategy, material for a breeding program and contribution to the development of coconut palms with a wide range of products.
2. Materials and methods
2.1 Plant materials
This research focused on the population of kopyor coconut in Ambulu, Wuluhan and Gumuk Mas Sub-districts in Jember Regency, East Java, using tall and dwarf types. Dwarf type consisted of green (GHK) and yellow fruits (GKK), while tall type included green (DHK), yellow (DKK) and brown fruits (DCK). The number of kopyor coconut palms in each sub-district is presented in .
Table 1. The number of kopyor coconut in each sub-district, Jember Regency.
2.2 Determination of the distribution of kopyor coconut palms
The distribution map of kopyor coconut palms in Jember Regency showed both tall and dwarf types. The area was determined by marking the coordinate position of each discovery, which was concentrated in a measuring plot unit GPS tagging (Global Positioning System). Furthermore, GIS was used to process GPS data collected at the area to generate vector graphics and enable the creation of distribution maps.
2.3 Phenotype characters of kopyor coconut palms
A purposive random sampling method was used to obtain kopyor coconut samples from farmers’ home gardens with a high concentration of at least 10-year-old trees producing fruit (Sukendah et al., Citation2011). The observed samples consisted of 10 palms from each type of kopyor coconut (GHK, GKK, DHK, DKK and DCK) found in the research area. The leaf samples having perfect openings were selected with female flowers that had opened completely, with a size of ±3 cm, and males measuring 3–5 mm. Additionally, kopyor coconut from dwarf and tall types were used in the fruit sample.
Various quantitative variables included (i) stem girth at 20 cm (cm), stem girth at 1.5 m (cm) and plant height 11 leaf scars (m), (ii) leaves with petiole length (cm), lamina length (cm), petiole thickness (cm), petiole width (cm), number of leaves, number of leaflets, leaf width (cm) and leaf length (cm), (iii) flowers with a length of peduncle (cm), inflorescence length (cm), peduncle width (cm), peduncle thickness (cm), number of female flowers, number of spikelets, spikelet length (cm) and the number of bunches. Kopyor fruit and the components were counted from three selected bunches per coconut palm and harvested 10–11 months after pollination. The variables observed were the number of fruit per bunch, the number of kopyor fruit per bunch, the percentage of kopyor fruit per bunch, the whole fruit weight (g), the polar fruit circumference (cm), the equatorial fruit circumference (cm), the polar fruit length (cm), the equatorial fruit length (cm), the fruit weight without husk (g), the fruit weight without husk and water (g).
The qualitative plant variables were (i) bole and cylindrical stems, (ii) a series of shoot and base leaves, (iii) inflorescence with female and male flowers, (iv) fruit and endosperm performance and (v) the shape of the tree, which included photographing the front view of coconut palm from each of GHK, GKK, DHK, DKK and DCK type.
2.4. Data analysis
The tools used were GPS, MapSource and coconut descriptor of the International Plant Genetic Resources Institute (IPGRI). The results of morphological observations were presented in tabular form and accompanied by images. The minimum, maximum and average values, as well as the coefficient of diversity, were calculated using the observations for GHK, GKK, DHK, DKK and DCK. Meanwhile, the diversity coefficient (DIVC) was calculated using the formula below (Singh & Chaudary, Citation1979):
SD = Standard deviation
xi = The ith data or ith value
= Average observed value
n = Total sample.
Analysis of variance was carried out on the combined response (index) of the observed variables. The combined response value obtained was the first principal component score value from the principal component analysis of the original variable response data (Abeyasekera, Citation2005). The Numerical Taxonomy and Multivariate System (NTSYS) version 2.02 was used to analyze morphological data and determine the degree of phenotypic similarity between GHK, GKK, DHK, DKK and DCK.
3. Results and discussion
3.1. Population of kopyor coconut palms
A survey conducted in Jember Regency identified 31 areas of concentrated kopyor coconut populations in three sub-districts, namely Wuluhan, Ambulu and Gumuk Mas, as shown in . Tall and dwarf kopyor coconuts were found in 20 and 11 different areas, respectively.
Figure 1. Map of kopyor coconut distribution in Wuluhan, Ambulu and Gumuk Mas sub-districts, Jember, East Java, Indonesia.
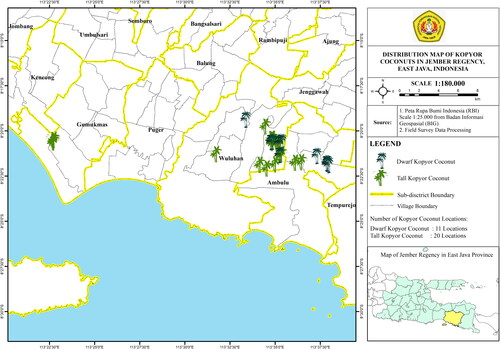
In Wuluhan, tall and dwarf types were found in 14 and 7 areas, respectively. The population of palms was reported in the villages of Kesilir, Ampel, Tegal Banteng and Babatan, with kopyor coconut growing in yards near residential areas. In Ambulu, there were 2 and 4 areas of tall and dwarf kopyor coconuts in Jati Gowok Village. Kopyor coconut was discovered in gardens near residential areas, where the population of tall type was more abundant. In Gumuk Mas, only tall type was found, and the population was spread over 4 areas in Mayangan Village in the maturation of fish ponds. According to the owner, tall type originated from Kesilir Village, Wuluhan Sub-District, due to the presence of several populations, as presented in .
3.2. Phenotype characters of kopyor coconut
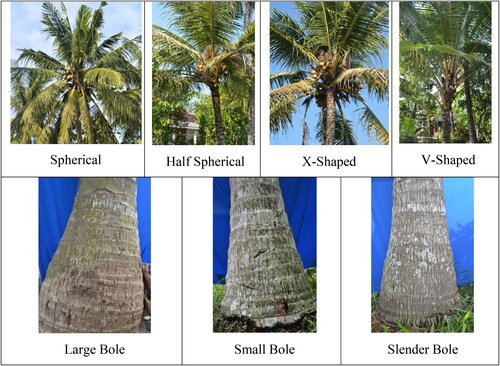
The morphological characters of kopyor coconuts in Jember based on crown shape, stem base, petiole and flower color and endosperm structure of kopyor showed quite high diversity. The heterogeneity of crown and stem base shape of kopyor coconut was presented in . In this context, four crown shapes were observed, spherical, half-spherical, X-shaped,and V-shaped. The spherical shape was commonly found among tall coconut populations, while the half-spherical dominated in dwarf type. In Macapuno population, the spherical shape was often found compared to the X-shape (Islam et al., Citation2013). The X-shape might be more common in highlands or seashores as a result of the adaptation process against strong winds.
The variation of stem base shape could be identified from the enlargement in the lowermost part of coconut where the roots were generally localized as bole. A total of three categories of bole shape were reported among kopyor coconut population namely, large, small and slender bole, as shown in . The research by Islam et al. (Citation2013) and Perera et al. (Citation2016) divided stem shapes into (1) no bole, (2) low or small bole and (3) high or large bole. Another classification proposed by Sugimura et al. (Citation1997) was slender, tapering and enlarged or bulbous bole. The stem base shape was one of the vegetative characteristics used to distinguish between dwarf and tall coconut. According to Perera et al. (Citation2016), dwarf type did not possess bole at the base of the stem. Kamaral et al. (Citation2017) also stated that dwarf coconut had small and short stature without a bole at the base. However, this research reported a bole trait presence in dwarf kopyor coconut. The results required more investigation to determine the specificity of the bole character of dwarf kopyor coconut in Jember.
Kopyor coconut in Jember had various petiole colors, namely green, yellow and brown, as shown in . Petiole color is a distinct trait used as a morphological marker in hybrid seedling identification (Arunachalam & Rajesh, Citation2008; Solangi et al., Citation2014). In coconut palm, the color of the petiole is equivalent to the stem. According to Rajesh (Citation2014), petiole color is the most widely used marker to select hybrid seedlings in the nursery stage. Even though the pigmentation does not improve yield or fruit quality, the color can be easily transmitted to the offspring after cross-pollination, serving as a convenient marker for the selection of hybrid (Islam et al., Citation2013).
Figure 3. The heterogeneity of shape and color of petiole, inflorescence, and fruit of kopyor coconut.
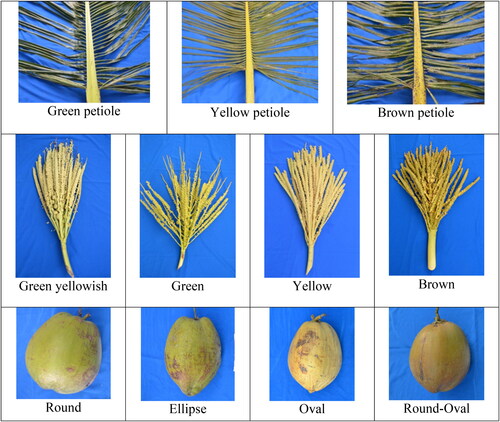
The variability of kopyor coconut inflorescence showed the same color as the flower stalks. For example, GKK was reddish yellow, with yellow flower stalks, while the green type was yellowish-green, accompanied by green flower stalks. The inflorescence color of DHK coconut was green similar to the stalk. Furthermore, the color of kopyor coconut fruits of all types was similar to the inflorescence. DKK coconut with inflorescence and fruit yellow color are examples. The results showed that the color diversity of inflorescence and kopyor coconut fruits was similar to the population of Pati, Central Java. Maskromo et al. (2015) reported a total of six variations of kopyor inflorescence and fruit color in dwarf types in Pati, which included green, brownish-green, brown, greenish-brown, yellow and orange. Aside from the inflorescence color, kopyor coconut had various fruit shapes, namely round, ellipse, oval, and round-oval.
Coconut endosperm exists in two forms, namely solid and liquid. In kopyor coconut, the solid endosperm passed through a mutation, changing into small particles which detached from the shell and mixed with the liquid endosperm to form a crumb structure. Previous research from Sumenep, East Java, showed that there was heterogeneity in endosperm crumb structure, classified into three types. Type A referred to endosperm separated into small clumps, which were detached from the shell, while Type B described endosperm, where some remained attached to the shell. Type C represented endosperm attached to the shell and relatively soft, containing a significant amount of coconut water (Sukendah et al., Citation2011).
This research showed heterogeneity in endosperm structure in dwarf and tall kopyor coconuts. The levels of endosperm structure were classified into four categories as shown in . Type A represented a soft-crumb structure with 76–100% endosperm detached from the shell, and little or no coconut water. Type B was characterized by a soft-crumb endosperm structure, where 51–75% were detached from the shell with coconut water. Type C, with a soft structure, comprised of 26–50% endosperm detached from the shell, and plenty of coconut water. Furthermore, Type D endosperm structure was soft with 0–25% detached from the shell, and there was plenty of coconut water.
Maskromo et al. (Citation2016) attempted to quantify the endosperm of kopyor structure using scoring levels. The quantity of dwarf type from Pati was classified into six scores, while Setiawan et al. (Citation2020) divided Kalianda tall kopyor coconut into nine scores. The endosperm was predicted to be controlled by a single recessive mutant k allele in a K locus (Sukendah et al., Citation2009) and the genotype was a homozygous kkk (Maskromo et al., Citation2016). Setiawan et al. (Citation2020) reported that the endosperm genotype of the high-quantity fruits was predicted as kkk/qqq, while the zygotic embryos were kk/qq. Additionally, the genotype was described by kkk resulting in a broken endosperm structure, and the high quantity was described by qqq. The fruits with low endosperm quantity were predicted as kkk/Q- -, including kkk/Qqq, kkk/QQq. K and Q loci were associated with kopyor trait and quantity of endosperm, respectively.
3.3. Phenotypic diversity and genetic relationship
3.3.1. Phenotypic diversity of kopyor coconut in Jember
Phenotypic diversity among kopyor coconut in Jember was examined in 11 vegetative and 17 reproductive characters. A wide range variability of stem and leaf characters was observed between tall and dwarf types, as shown in . Trunk size heterogeneity was observed, allowing for significant distinction, where the plant height of tall types was twice to three times dwarf. Tall types had an average plant height of 11.75 m, while dwarf was 5.28 m.
Table 2. Average, standard deviation, diversity of coefficient and minimum–maximum value of kopyor coconut.
The results showed that bole, stem girth, and plant height in dwarf kopyor coconut population had a DIVC higher than in tall type, as shown in . Dwarf kopyor coconut had a larger bole and stem circumference than tall type. This phenomenon is likely caused by the dwarf kopyor coconut in Jember growing among tall type, resulting in hybrid kopyor coconuts.
Morphology of tall kopyor coconut leaves had a longer petiole and lamina length than dwarf type, resulting in a greater number of leaflets, as shown in . However, the DIVC of leaf width and number of leaves in tall kopyor coconut was less than in dwarf types. Meanwhile, tall population showed less diversity compared to dwarf type in terms of vegetative characteristics.
In the context of reproductive characters, the highest variation was observed in the number of female flowers, which was the DIVC for dwarf and tall types at 46.12% and 33.03%, respectively. The ability of each coconut palm to produce female flowers depends on the genetic potential of individual plants. Therefore, the high numbers in each bunch increase the possibility of fruit formation after pollination (Maskromo et al., Citation2016). In this research, the numbers of fruits per bunch were 5.55 and 6.45 for dwarf and tall kopyor coconuts with percentages of 29.0% and 28.0%, respectively. The results were slightly lower than Pati dwarf type which had a percentage ranging from 24.8% to 38.9% (Maskromo et al., Citation2013). Besides genetic potential, the environment such as rainfall also influences the production of female flowers and fruit formation. shows that the research area experienced significant rainfall a year before pollination, ranging from 1275 to 1632 mm per year. The availability of sufficient water influenced the success of the initiation and formation of female flowers and kopyor coconut inflorescences. These patterns also affected the formation and development, with rainfall ranging from 600 to 967 mm. Reduced rainfall during fruit formation had a positive impact, specifically in reducing falls.
Table 3. Rainfall data in 2018 to 2019 in Jember.
Among the fruit characters, the highest variation of 59.88% and 34.20% was found in the number of kopyor nuts per bunch in dwarf and tall types, followed by the percentage of fruit, number of bunches and endosperm weight. This high diversity of fruit component characters was due to outcrossing pollination among the various types. According to Larekeng et al. (Citation2018), the spread of kopyor coconut pollen from Pati reached 54 m and the occurrence of outcrossing frequency was 95%.
High diversity in the vegetative and reproductive characteristics such as stem girth, plant height, leaf width, number of leaves, petiole length, number of female flowers, number of kopyor fruit, percentage of kopyor fruit per bunch and endosperm weight was used for dwarf kopyor coconut improvement. The results were consistent with Miftahorrachman et al. (Citation2017), where the number of leaves, number of leaflets, plant height, stem circumference, number of bunches, petiole length, petiole width, rachis length and leaf length were used as selection characters to increase production. Similarly, in sweet endosperm with mutant characteristics resembling kopyor coconut population from Maharashtra, India, the highest variation was detected in plant height, number of leaves, number of female flowers, percentage of sweet kernel fruits and number of nuts per bunch (Samsudeen et al., Citation2013). The high phenotypic and genetic coefficient variation of the characters was also found in normal coconut (Suchithra & Paramaguru, Citation2018).
Recent research has used molecular markers as a tool for the selection of crop improvement. However, the selection based on phenotypic characters is the main technique used in breeding programs, specifically in coconut. Besides vegetative characters, high heterogeneity in reproductive characteristics such as the number of kopyor fruit per bunch, percentage of kopyor fruit and endosperm weight can be used as a selection character for the breeding programs. Natarajan et al. (Citation2010) stated that the number and length of leaves as well as petioles showed a positive correlation with nuts yield. According to Harries (Citation1978), fruit component characters are significantly important in classifying coconut types.
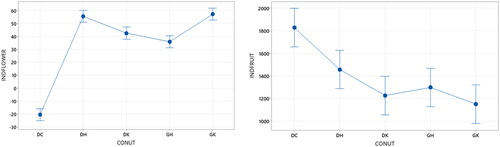
Trait indices comparison by combined response analysis in dwarf kopyor coconut populations showed differences between yellow and green types based on the stem, leaf, and flower traits, except for fruit traits, as shown in . Yellow dwarf kopyor (GKK) had a higher stem and flower index value than green (GHK). However, the leaf index was lower and GKK fruit index value was not different from GHK. In tall kopyor coconut, the stem index value of DKK was lower than DHK and DCK. Meanwhile, DHK stem index value was not different from DCK. The leaf and flower index values of DKK, DHK and DCK were different and DCK was the smallest. However, DCK fruit index value was the largest, as shown in .
Table 4. Trait indices in and between dwarf and tall kopyor coconuts.
A comparison of dwarf and tall kopyor coconuts showed that the stem index values of GKK and GHK were not different from DHK and DCK. In this context, the flower index values of GKK and GHK were higher than DCK. On the contrary, the fruit index values of GKK and GHK were lower than DCK. The fruit index values of GKK, GHK, DKK and DHK were the same or there was no difference. The combined response analysis of the various morphological characters observed can be used as a differentiating factor between similar and different types of kopyor coconut.
3.3.2. Genetic relationship of kopyor coconut in jember
Further analysis of the genetic relationship of kopyor coconut populations was conducted based on morphological character. The phylogenetic analysis categorized the type into three clusters, as shown in . Cluster I consisted of tall kopyor coconut (DHK, DCK and DKK), Cluster II comprised a yellow dwarf, and Cluster III included a green dwarf. The similarity between DHK kopyor coconut, DCK and DKK was 24% but the value was 29% between the DCK and DKK. In addition, GKK and GHK had a 6% similarity among dwarf kopyor coconut.
Figure 6. Phylogenetic relationship tree based on morphological character of kopyor coconut. GKK: yellow dwarf kopyor; GHK: green dwarf kopyor; DHK: green tall kopyor; DKK: yellow tall kopyor; DCK: brown tall kopyor.
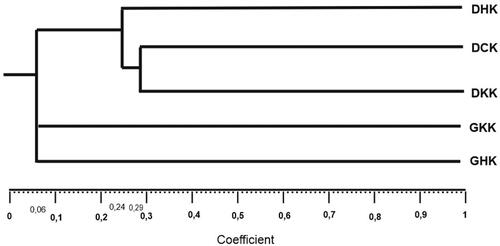
The similarity among dwarf kopyor coconut was less than tall type. Therefore, dwarf kopyor coconut was more diverse than tall type, and this was supported by the quantitative evidence in . Several research showed the same results as reported by Miftahorrachman et al. (Citation2017), where GKK coconut in Pati showed the highest diversity in growth character. Phylogenetic tree based on SNAP and InDel markers placed tall and dwarf kopyor coconut in one cluster, while tall type formed a separate sub-group with a greater similarity value than dwarf (Rahmawati et al., Citation2022). In Puan Kalianda, tall type showed 40% similarity based on SSR markers (Rahayu et al., Citation2021). In Jember, the type was placed in one cluster with Sumenep, Banten and Kalianda based on genotype data, as calculated using a dissimilarity matrix (Pesik et al., Citation2017). The greater the similarity between the characters, the higher the value, increasing the closeness of the phylogenetic relationship (Fatimah, Citation2013). Julisaniah et al. (Citation2008) stated that the chance of success decreased with increased phylogenetic relationship. However, there was an increased possibility of obtaining a superior genotype when a successful cross was achieved.
4. Conclusion
In conclusion, kopyor coconuts in Jember were reported to show high variability with similarities ranging from 6% to 29% based on phenotypic characters. The palm population was dominated by tall kopyor coconut which was often found in house yards in Wuluhan, Ambulu and Gumuk Mas. Meanwhile, generative characteristics such as the number of female flowers, as well as the number and percentage of kopyor fruits per bunch were the most diverse morphological characteristics used for the selection of breeding programs. Among the population, DCK had the smallest and largest flower and fruit index values, respectively. In this context, the results were useful for kopyor coconut improvement programs. The diversity of dwarf and tall kopyor coconuts in Jember could be explored for the formation of hybrid types specific to Jember ecotype.
Acknowledgments
The authors are grateful to kopyor coconut farmers in Wuluhan who assisted in collecting data as well as the Ministry of Education, Culture, Research and Technology for providing research funding.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abeyasekera, S. (2005). Multivariate methods for index construction.Chapter XVIII: Household sample surveys in developing and transition countries (pp. 1–12). https://mdgs.un.org/unsd/hhsurveys/pdf/Chapter_18.pdf.
- Amzeri, A., Indradewa, D., Daryono, B. S., & Rachmawati, D. (2011). Family of madurese maize (Zea mays L.) based on morphological characters and RAPD markers. Biota: Jurnal Ilmiah Ilmu-Ilmu Hayati, 16(2), 227–235. https://doi.org/10.24002/biota.v16i2.104
- Arunachalam, V., & Rajesh, M. K. (2008). Breeding of coconut palm (Cocos nucifera L.). CABI Reviews, 3 (053), 1–12. https://doi.org/10.1079/PAVSNNR20083053
- Chomchalow, N. (1999). Coconut varieties in Thailand. AU Journal of Technology, 3(1), 19–30. https://repository.au.edu/server/api/core/bitstreams/b601e0b3-15b0-4f74-a99e-43214fe6d3cf/content
- Fatimah, S. (2013). Morphological analysis and relationship of eleven species of Salak Plants (Salacca zalacca (Gertner) Voss) Bangkalan. Agrovigor, 6(1), 1–11. https://journal.trunojoyo.ac.id/agrovigor/article/viewFile/1460/1250
- Gunathilake, K., & Samarasinghe, D. (2015). Evaluation of properties of Dikiri Pulp in the formulation of Jam. Cord, 31(2), 26–32. https://doi.org/10.37833/cord.v31i2.59
- Harries, H. C. (1978). The evolution, dissemination and classification of Cocos nucifera L. The Botanical Review, 44(3), 265–319. https://doi.org/10.1007/BF02957852
- Islam, M. N., Azad, A. K., Namuco, L. O., Borromeo, T. H., Cedo, M. L O., & Aguilar, E. A. (2013). Morphometric characterization and diversity analysis of a makapuno coconut population in UP Los banos. Pakistan Journal of Agricultural Research, 26(4), 254–264. https://www.academia.edu/100386753
- Jerard, B. A., Damodaran, V., Niral, V., Samsudeen, K., Rajesh, M. K., & Sankaran, M. (2013). Conservation and utilization of soft endosperm coconut accession from Andaman Islands. Journal of Plantation Crops, 41(1), 14–21. https://www.researchgate.net/profile/Rajesh-Mk/publication/247792193
- Jerard, B. A., Niral, V., Samsudeen, K., Nair, R. V., Jayabose, C., & Thomas, G. V. (2015). Development of a dwarf x tall coconut hybrid ‘Kalpa Samrudhi’. Journal of Plantation Crops, 43(1), 46–52. http://krishi.icar.gov.in/jspui/handle/123456789/24124
- Julisaniah, N. I., Sulistyowati, L., & Sugiharto, A. N. (2008). Relationship Analysis of Cucumber (Cucumis sativus L.) Using RAPD-PCR and Isozyme Methods. Biodiversitas Journal of Biological Diversity, 9(2), 99–102. https://doi.org/10.13057/biodiv/d090205
- Kamaral, L. C. J., Perera, S., Perera, K., & Dassanayaka, P. N. (2017). Characterization of Sri Lanka yellow dwarf coconut (Cocos nucifera L.) by DNA fingerprinting with SSR markers. Journal of the National Science Foundation of Sri Lanka, 45(4), 405.https://doi.org/10.4038/jnsfsr.v45i4.8234
- Kurniasih, S., Rubiyo, Setiawan, A., Purwantara, A., & Sudarsono, . (2020). Analysis of genetic diversity of cocoa (Theobroma cacao L.) germplasm based on SSR markers. Jurnal Penelitian Tanaman Industri, 17(4), 156–162.https://doi.org/10.21082/jlittri.v17n4.2011.156-162
- Larekeng, S. H., Purwito, A., Mattjik, N. A., & Sudarsono, S. (2018). Microsatellite and SNAP markers used for evaluating pollen dispersal on pati tall coconuts and xenia effect on the production of ‘Kopyor’ fruits. IOP Conference Series: Earth and Environmental Science, 157(1), 012042. 1315/157/1/012042 https://doi.org/10.1088/1755-1315/157/1/012042
- Manimekalai, R., & Nagarajan, P. (2010). Bulk line analysis in coconut (Cocos nucifera L.) for inferring relationship between talls, dwarfs and niu Leka Dwarf forms. Indian Journal of Plant Genetic Resources, 23(1), 77–81.
- Maskromo, I., . Novarianto, H., Sukendah, . Sukma,D., & Sudarsono. 2013. Productivity of three Dwarf Kopyor coconut varieties from Pati, Central Java, Indonesia. Coconut Research and Development (Cord), 29(2), 10. https://doi.org/10.37833/cord.v29i2.86
- Maskromo, I., Novarianto H.,Sukendah, & Sukma, D., & Sudarsono. (2014). Diversity of the fruit phenotypes and endosperm quality of Kalianda Tall Kopyor and Pati Dwarf Kopyor coconuts. Bul Palma, 15, 102–109. https://doi.org/10.21082/bp.v15n2.2014.102-109
- Maskromo, I., Tenda, E. T., Tulalo, M. A., Novarianto, H., Sukma, D. ., Sukendah, ., & Sudarsono . (2016). Phenotypic and genetic diversity of three Dwarf Kopyor coconuts from Pati, Central Java. Jurnal Penelitian Tanaman Industri, 21(1), 1. https://doi.org/10.21082/littri.v21n1.2015.1-8
- Maskromo, I., Larekeng, S.H., Novarianto, H., & Sudarsono, . (2016). Xenia negatively affecting Kopyor nut yield in Kalianda Tall Kopyor and Pati Dwarf Kopyor coconuts. Emirates Journal of Food and Agriculture, 28(9), 644–652. https://doi.org/10.9755/ejfa.2015-07-552
- Miftahorrachman, Mawardi, S., & Novarianto, H. 2017. Correlation and path coefficient analysis of Kopyor Dwarf coconut (Cocos nucifera L.). Coconut Research and Development (Cord), 33 (1):15. https://doi.org/10.37833/cord.v33i1.51
- Natarajan, C., Ganesamurthy, K., & Kavitha, M. (2010). Genetic variability in coconut (Cocos nucifera). Electronic Journal of Plant Breeding, 1(5), 1367–1370. https://core.ac.uk/download/25767353.pdf
- Novarianto, H., Maskromo, I., & Sudarsono,. 2014. Production technology for Kopyor coconut seed nuts and seedlings in Indonesia. Coconut Research and Development (Cord), 30(2), 10. https://doi.org/10.37833/cord.v30i2.77
- Perera, L., Baudouin, L., & Mackay, I. (2016). SSR markers indicate a common origin of self-pollinating Dwarf coconut in South-East Asia under domestication. Scientia Horticulturae. 211, 255–262. https://doi.org/10.1016/j.scienta.2016.08.028
- Pesik, A., Efendi, D., Novarianto, H., Dinarti, D., & Sudarsono, S. (2017). Development of SNAP markers based on nucleotide variability of WRKY genes in coconut and their validation using multiplex PCR. Biodiversitas Journal of Biological Diversity, 18(2), 465–475. https://doi.org/10.13057/biodiv/d180204
- Rahmawati, A., Dinarti, D. ., Maskromo, I., Volkaert, H. A., & Sudarsono,. (2022). Coconut diversity based on chloroplast single nucleotide amplified polymorphism (SNAP) and insertion-deletion (InDel) markers. Biodiversitas Journal of Biological Diversity, 23(8), 4073–4081. https://doi.org/10.13057/biodiv/d230827
- Rahayu, M. S., Setiawan, A., Maskromo, I., Purwito, A., & Sudarsono, . (2021). Genetic diversity analysis of Puan Kalianda Kopyor coconuts (Cocos nucifera) from South Lampung, Indonesia based on SSR markers. Biodiversitas Journal of Biological Diversity, 23(1), 205–211. https://doi.org/10.13057/biodiv/d230126
- Rajesh, M. K., Jerard , B.A, Preethi, P., Thomas, R. J., & Karun, A. (2014). Application of RAPD markers in hybrid verification in coconut. Crop Breeding and Applied Biotechnology, 14(1), 36–41. https://doi.org/10.1590/S1984-70332014000100006
- Toan, N. B., & Thanh, V. C. (2011). Potential development of makapuno coconut in the mekong delta of Vietnam. International Journal of Renewable Energy, 6, 21–28. https://doi.org/10.14456/iire.2011.10
- Samsudeen, K., Rajesh, M. K., Nagwaker, D. D., Reshmi, R., Kumar, P. A., Devadas, K., & Anitha, K. (2013). Diversity in Mohachao Narel, a sweet endosperm coconut (Cocos nucifera L.) population from Maharashtra, India. National Academy Science Letters, 36(3), 319–330. https://doi.org/10.1007/s40009-013-0128-0
- Setiawan, A., Rahayu, M. S., Maskromo, I., Purwito, A., & Sudarsono .,. 2020. Inheritance pattern of endosperm quantity and Kopyor coconut (Cocos nucifera L.) fruit variations. IOP Conference Series: Earth and Environmental Science, 418(1), 012039. https://doi.org/10.1088/1755-1315/418/1/012039
- Singh, R. K., & Chaudary, B. D. (1979). Biometrical methods in quantitaive genetics analysis. Kaylani Publisher.
- Sivakumar, V., Sudha, R., Niral, V., Neema, M., & Geethanjali, S. (2022). Makapuno-A gelatinous mutant coconut variety. Vigyan Varta, 3(9), 15–20.
- Solangi, A. H., Baloch, P. A., Siddiqui, A. A., Uddin, R., Nizamani, F. K., & Iqbal, M. Z. (2014). Seedling growth of three coconut (Cocos nucifera L.) varieties in Karachi, Pakistan. International Journal of Biological Research, 2(2), 153–154. https://ijbr.net/journals/vol22/15-14-022-Short-Communication-Abdul-Hameed-Solangi.pdf
- Suchithra, M., & Paramaguru, P. (2018). Variability and correlation studies for vegetative, floral, nut and yield characters in indigenous and exotic coconut genotypes. International Journal of Current Microbiology and Applied Sciences, 7(07), 3040–3054. https://doi.org/10.20546/ijcmas.2018.707.35
- Sugimura, Y., Itano, M., Salud, C. D., Otsuji, K., & Yamaguchi, H. (1997). Biometric analysis of diversity of coconut palm: Cultivar classification by botanical and agronomic traits. Euphytica, 98(1–2), 29–35. https://doi.org/10.1023/A:1003053128120
- Sukendah, Volkaert, H., & Sudarsono. (2009). Isolation and analysis of DNA fragment of genes related to Kopyor trait in coconut plant. Indonesian Journal of Biotechnology, 14(2), 1169–1178. https://doi.org/10.22146/ijbiotech.7814
- Sukendah, Abidin, Z., Wiyatiningsih, S., & Wijayanti, B. W.(2011). Morphological characters of Kopyor coconut grown in Sumenep, Madura, Indonesia. Isnar C2FS Proceeding (pp. 245–254). Surabaya, June 27-28, 2011. Faculty of Agriculture, Universitas Pembangunan Nasional "Veteran" Jawa Timur. ISBN: 9786028915939.
- Swaminathan, M. S., & Nambiar, M. C. (1961). Cytology and origin of the Dwarf coconut palm. Nature, 192(4797), 85–86. https://doi.org/10.1038/192085a0
- Wicaksono, A., Raihandhany, R., & da Silva, J. A T. (2021). Kopyor versus macapuno coconuts: are these two edible mutants of Southeast Asia the same? Planta, 254(5), 86. https://doi.org/10.1007/s00425-021-03740-y
- Xiao, Y., Xu, P., Fan, H., Baudouin, L., Xia, W., Bocs, S., Xu, J., Li, Q., Guo, A., Zhou, L., Li, J., Wu, Y., Ma, Z., Armero, A., Issali, A. E., Liu, N., Peng, M., & Yang, Y. (2017). The genome draft of coconut (Cocos nucifera). GigaScience, 6(11), 1–11. https://doi.org/10.1093/gigascience/gix095