ABSTRACT
Cisplatin is the commonly used chemotherapeutic drug in treatment of various cancers. However, development of resistance towards cisplatin results in tumor recurrence. Here, we aim to understand the mechanisms of action of cisplatin and emergence of resistance to cisplatin using mass spectrometry-based proteomic approach. A panel of head and neck squamous cell carcinoma (HNSCC) cell lines were treated with cisplatin at respective IC50 for 24 h and label-free mass spectrometry analysis was carried out. Proteomic analysis of A253, FaDu, Det562 and CAL27 cell lines upon cisplatin treatment resulted in the identification of 5,060, 4,816, 4,537 and 4,142 proteins, respectively. Bioinformatics analysis of differentially regulated proteins revealed proteins implicated in DNA damage bypass pathway, translation and mRNA splicing to be enriched. Further, proteins associated with cisplatin resistance exhibited alterations following short-term cisplatin exposure. Among these, class III tubullin protein (TUBB3) was found to be upregulated in cisplatin-treated cells compared to untreated cells. Western blot analysis confirmed the elevated expression of TUBB3 in cells treated with cisplatin for 24 h, and also in cisplatin resistant HNSCC cell lines. This study delineates the early signaling events that enable HNSCC cells to counteract the cytotoxic effects of cisplatin and facilitate the development of resistance.
Introduction
The discovery of anticancer properties of cisplatin was a breakthrough in tumor treatment. Cisplatin was the first platinum drug to be approved by the FDA as early as 1978 in treatment of testicular , ovarian and bladder cancer.Citation1 Platinum-based drugs including cisplatin, carboplatin, and oxaliplatin still remains a backbone in treatment of various cancers. Cisplatin continues to be choice of drug in the treatment of 20 different tumor types including head and neck cancer, lung cancer, and ovarian cancers.Citation2 Primary target of cisplatin is DNA and binding of cisplatin to DNA results in the formation of DNA adducts. This further activates DNA damage response and apoptotic pathways eventually leading to cell death. Although cisplatin induced molecular aberrations in cells are studied, the activation of specific signaling pathways upon cisplatin treatment are not completely understood, especially the early signaling events. Identifying these early molecular changes will provide clue on the factors that determine the fate of damaged cell toward apoptosis or cell cycle.Citation3 Understanding these signaling pathways will further pave way to delineate the mechanisms of drug resistance employed by cells and to develop a targeted therapy.
Towards this, Ebhardt HA, et al., employed a targeted mass spectrometry-based proteomic approach to determine the immediate response of pancreatic cancer cells to six drugs and their combinations used in pancreatic cancer treatment.Citation4 This study showed that the downregulated proteins Jupiter microtubule associated homolog 1 (JPT1), p21 (RAC1) activated kinase 1 (PAK1) and sperm associated antigen 5 (SPAG5) could serve as early indicators of drug efficacy, whereas the upregulated proteins, 14-3-3 and kallikrein related peptidase 2 (KLK2) play a role as potential early drug response markers and could be used in targeted therapy. Musiani D, et al., carried out proteomic analysis of ovarian cancer cells treated with cisplatin for 6 and 24 h. Pim-2 proto-oncogene, serine/threonine kinase (PIM2), a kinase was found to be upregulated in cells treated with cisplatin and overexpression of PIM2 was able to rescue cells from undergoing apoptosis via regulation of BAD. This study has shown that PIM2 to be involved in regulating apoptosis, cell survival, and DNA damage in response to cisplatin, thus proving that PIM2 can be used as druggable target to improve platinum sensitivity.Citation5 In a recent study, the authors employed proteomics as a tool to understand the mechanisms of action of over 800 compounds by examining the changes in proteome of HCT116 cell line upon 24 h treatment.Citation6 Mitchell et. al. and Frenjo et. al. recently published an activity landscape of 125 cell lines from NCI60 and CRC65 cohort analyzing early proteomics response to over more than 200 drugs identifying markers of drug sensitivity, resistance, and common mode of action.Citation7,Citation8 However, these repositories lack information pertaining to HNSCC.
In India, HNSCC is one of the top-most common cancers and oral squamous cell carcinoma (OSCC), a sub-type of HNSCC is the 2nd most common cancer.Citation9 Alcohol consumption, tobacco use (both smoke and smokeless form), and infection by human papilloma virus (HPV) are the major risk factors in the development of oral cancer. Surgery, radiation therapy and chemotherapy are the treatment options available for oral cancer patients. Cisplatin (cis-diaminedichloroplatinum, CDDP) is the most commonly used chemotherapeutic drug for OSCC. However, development of chemoresistance either inherited or acquired has limited the use and efficacy of cisplatin in oral cancer treatment. Several studies have been carried out to understand the mechanisms of cisplatin resistance in head and neck cancer.Citation1,Citation10 However, there are only a few studies reporting the immediate response of cells to cisplatin. In this study, we employed label-free quantitative mass spectrometry-based approach to understand the immediate protein alterations that occur upon cisplatin treatment. We could identify several proteins to be altered in a panel of HNSCC cell lines upon cisplatin treatment for 24 h. Further, one of the upregulated proteins was validated by Western blot analysis not only in cells exposed to cisplatin but also in cisplatin resistant cells.
Materials and methods
Cell culture conditions
Four HNSCC cell lines, A253, FaDu, Det562, and CAL27 were used in this study. All these cell lines were from ATCC. A253 is derived from submaxillary salivary gland of an epidermoid carcinoma patient. A253 cells were cultured in MaCoy’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin mixture. FaDu, a squamous cell carcinoma cell line derived from a patient with hypopharyngeal tumor was grown and maintained in Roswell Park Memorial Institute (RPMI) medium containing 10% fetal bovine serum and 1% penicillin/streptomycin mixture. Det562 is a pharyngeal carcinoma cell line and CAL27 is a squamous cell carcinoma cell line derived from tongue. These two cell lines were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin mixture. All these cells were grown at 37°C in a humidified 5% CO2 incubator.
Sensitivity of HNSCC cells to cisplatin
Cisplatin used in the study was purchased from Selleckchem Chemicals (catalog #: S1166) and cisplatin stock was prepared as per the manufacture details. HNSCC cells were seeded at a density of 3 × 103 cells per well in a 96-well plate. Post-24 h, cells were treated with varying concentration of cisplatin ranging from 1–8 µM for 72 h. Viability of cells was further determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The absorbance was measured at 570 and 650 nm using a Microplate Spectrophotometer (Multiskan GO, Thermo Scientific, Bremen, Germany).
Sample preparation for mass spectrometry analysis
HNSCC cells were grown to 70–80% confluency. Once they reached the required confluency, cells were treated with cisplatin at their respective IC50 for 24 h. Untreated cells served as control. Post-treatment, cells from both the conditions were washed with 1X PBS thrice and harvested in 2% SDS buffer containing protease and phosphatase inhibitors. The samples were then sonicated, centrifuged and protein concentration was determined using bicinchoninic acid (BCA) assay. Equal amount of protein lysate from untreated and cisplatin treated condition was subjected to reduction, alkylation and trypsin digestion as described previously.Citation11 The peptides were then subjected to label-free quantification using mass spectrometry.
LC-MS/MS analysis
The dried peptides were reconstituted 0.1% formic acid before injecting into the mass spectrometer. Data was acquired on the Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, Bermen, Germany) coupled with Easy n-LC 1000 liquid chromatography system (Thermo Fisher Scientific). Data for Det562 cell line pair was acquired on Q Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with Dionex Ultimate 3000 HPLC system. Data were acquired in triplicates. Samples were loaded on to a two-column setup, the peptides were first loaded onto the trap column at a flow rate of 6 ul/mins, followed by separation in the analytical column with a gradient of 8–38% solvent B (96% Acetonitrile in 0.1% Formic acid for Easy nLC 1000 and 80% Acetonitrile in 0.1% Formic acid for Dionex Ultimate 3000) over 165 minutes at a flow rate of 280 nl/min with a total run time of 180 minutes. The peptides were analyzed on an orbitrap in DDA mode with a cycle time of 3 seconds. Resolution at 200 m/z was 120,000 for full scan and 30,000 for MS2. HCD was used for fragmentation of the selected parent ions at 34% NCE (Normalized Collision Energy). Dynamic exclusion was set to 30 seconds. AGC target was set to 4e5 for full scan and 1e5 for MS2 with maximum injection time of 50 ms and 54 ms, respectively.
Database searches for peptide and protein identification
Data from LC-MS/MS analysis was searched using Proteome Discoverer (version 2.2, Thermo Scientific, Bremen, Germany) suite against NCBI RefSeq database (release 89 with known contaminants). We used SequestHT and MS Amanda as search engines. The search parameters included acetylation on N-terminal of protein; oxidation on methionine as dynamic modification, and carboxyamidomethylation on cysteine as static modification. The enzyme used to generate peptides was trypsin with a maximum of two missed cleavages. Precursor mass tolerance of 10 ppm and fragment mass tolerance of ±0.05 Da was applied. FDR of 1% was applied at the peptide spectrum match (PSM), peptide and protein level. Minora feature detector node was used with feature mapper for label free quantification of peptides and proteins.
Availability of data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDECitation12 partner repository with the dataset identifier PXD045610.
Bioinformatics analysis
The pathway enrichment analysis of the differentially regulated proteins was performed using Reactome Pathway database using the ClueGo (version 2.5.8) plug-in in Cytoscape (version 3.9.1) software.Citation13,Citation14 The protein-protein interaction network was constructed using the software Pathvisio.Citation15
Western blot analysis
CAL27 and Det562 cells were treated with cisplatin at IC50 for 24 h and harvested in RIPA buffer containing protease and phosphatase inhibitors. Similarly, untreated and cisplatin resistant CAL27 and Det562 cells were cultured to 70% confluence and harvested in RIPA lysis buffer. Western blot analysis was carried out as described previouslyCitation11 and was repeated thrice. Antibody to TUBB3 was purchased from Abcam (Catalog #: ab231083). β-actin antibody used as loading control was purchased from Sigma (Catalog #: A5316).
Cellular proliferation assay
Cisplatin resistant and control CAL27 and Det562 cells were seeded at a density of 3 × 10Citation3 cells per well in triplicate in 96-well plates. Cellular proliferation was monitored for a period of 6 days. The proliferation rate of cells was determined by MTT assay as described above at different time intervals, day 2, day 4, and day 6.
Results
Determination of half-maximum inhibitory concentration (IC50) of HNSCC cell lines
HNSCC cell lines, A253, CAL27, Det562, and FaDu were treated with varying concentrations of cisplatin and their IC50 was determined by MTT assay. As shown in , the IC50 for A253, FaDu, Det562, and CAL27 was found to be 1.5 µM, 2 µM, 1.5 µM, and 2 µM, respectively. Further, to understand the early molecular changes induced in response to cisplatin treatment, mass spectrometry-based label-free quantitative (LFQ) proteomic approach was employed.
Figure 1. (a) Response of HNSCC cell lines, A253, CAL27, Det562 and FaDu to cisplatin: HNSCC cells were treated with varying concentration of cisplatin for 72 h and IC50 was determined by MTT assay. The IC50 of A253, FaDu, Det562 and CAL27 was found to be 1.5 µM, 2 µM, 1.5 µM and 2 µM, respectively (b) Workflow employed to identify proteins altered in response to cisplatin. HNSCC cells were treated with cisplatin at their IC50 for 24 h. Untreated cells served as control. Equal amount of protein from both the conditions was subjected to trypsin digestion followed by mass spectrometry analysis.
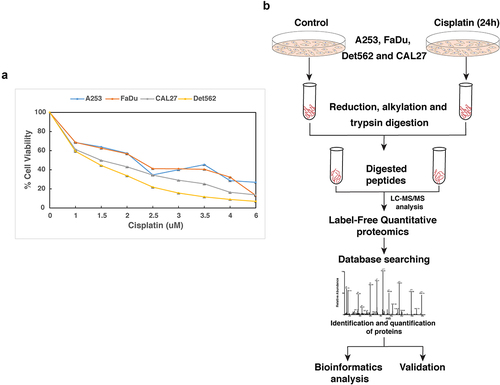
Mass spectrometry-based analysis of head and neck cancer cells treated with cisplatin
Studies have shown that a 24 h time point would be an appropriate time point to profile the perturbed proteome in response to a pharmacological agent and to elucidate the mechanisms of action of several drugs.Citation4,Citation6 Cisplatin is shown to induce migration and invasion of cancer cells and expression of several proteins implicated in cisplatin resistance following 24 h treatment.Citation16,Citation17 A comparative phosphoproteomic analysis of ovarian cancer cells treated with cisplatin for 6 h and 24 h, showed an enhancement of signaling events at 24 h treatment compared to 6 h treatment duration.Citation5 Thus, in this study, we choose an 24 h timepoint to understand the early proteomic changes that occur upon cisplatin treatment. HNSCC cell lines were grown to 70% confluency and treated with cisplatin at IC50 for 24 h. Following 24 h treatment, cells from both the conditions were harvested and processed for mass spectrometry analysis as shown in . LC-MS/MS analysis led to the identification of 5,060, 4,816, 4,537, and 4,142 proteins in A253, FaDu, Det562, and CAL27, respectively. Among these, 230 proteins were upregulated and 122 proteins were found to be downregulated in cisplatin treated cells compared to untreated cells (with high confidence and 2-fold cutoff). Complete list of proteins identified in the study is provided in Supplementary Table S1. Partial list of differentially regulated proteins is provided in .
Table 1. Partial list of proteins that are differentially regulated in cisplatin treated HNSCC cell lines compared to untreated cells.
Effects of cisplatin on DNA
DNA is the primary target of cisplatin and binding of cisplatin results in the formation of inter- and intra-strand crosslinks. To overcome these damages, cells activate DNA damage repair pathways such as nucleotide excision repair pathway, trans lesion polymerase, and homologous recombination.Citation18 In this study, approximately 60 proteins associated with DNA damage response and repair exhibited altered expression levels following cisplatin treatment. A list of some of these proteins is provided in . The protein-protein interaction (PPI) network for these proteins was manually constructed based on evidence from published literature using the tool, Pathvisio. illustrates the resulting protein-protein interaction network. Among 64 proteins implicated in DNA damage response and repair, around 50 proteins were found to interact in the PPI network.
Figure 2. Protein-protein interaction (PPI) network of proteins implicated in DNA damage response and repair. Proteins associated with DNA damage and repair were shortlisted from list of proteins altered upon cisplatin treatment. An PPI network was created based on literature evidence using the tool, Pathvisio. The number on each protein indicates the fold change observed upon cisplatin treatment compared to untreated cells.
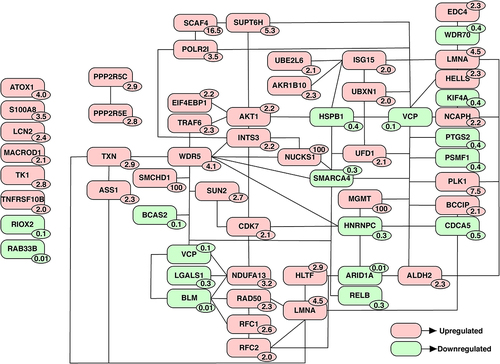
Table 2. Partial list of proteins involved in DNA damage response and repair and are differentially expressed upon cisplatin treatment.
Pathway analysis of differentially expressed proteins
A pathway-based enrichment analysis of differentially regulated proteins was carried using Reactome pathway database in Cytoscape software. In addition to proteins implicated in DNA damage bypass pathway, proteins involved in translation and mRNA splicing were found to be enriched (). Some of the proteins implicated in DNA damage bypass pathway include Ubiquitin conjugating enzyme E2 L6 (UBE2L6), ISG15 ubiquitin like modifier (ISG15) and DNA polymerase epsilon 4, accessory subunit (POLE4), replication factor C subunit 1 (RFC1), and replication factor C subunit 2 (RFC2) were found to be upregulated in this study. Several proteins reported to play a role in mRNA translation were enriched in response to cisplatin treatment. The expression of several ribosomal proteins, ribosomal protein S9 (RPS9) ribosomal protein S24 (RPS24) ribosomal protein L15 (RPL15), ribosomal protein L11 (RPL11), ribosomal protein L19 (RPL19), and ribosomal protein L29 (RPL29) involved in translation were also found to be altered in cisplatin treated HNSCC cell lines.
Figure 3. Pathway enrichment analysis of proteins altered in response to cisplatin treatment was carried out using ClueGo plugin in Cytoscape. This analysis revealed enrichment of proteins implicated translation, mRNA splicing and DNA damage bypass pathways among others to be enriched upon cisplatin treatment. Proteins highlighted in red indicates upregulation and downregulated proteins are in green.
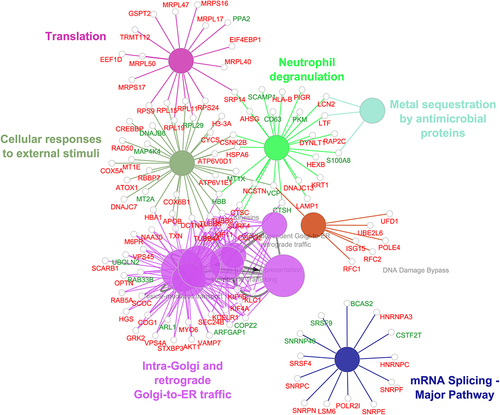
Further, proteins associated with mRNA splicing were found to be altered in response to cisplatin treatment. Heterogeneous nuclear ribonucleoprotein C (HNRNPC), heterogeneous nuclear ribonucleoprotein A3 (HNRNPA3), BCAS2 pre-mRNA processing factor (BCAS2), small nuclear ribonucleoprotein polypeptides, SNRPC, SNRPN, SNRPF, and SNRPE and serine and arginine rich splicing factors SRSF9 and SRSF4 were enriched. Heterogeneous nuclear ribonucleoproteins, HNRNPC and HNRNPA3 have been previously reported to be overexpressed in cisplatin resistant cell lines.Citation19,Citation20 It has been shown that HNRNPC overexpression promotes chemoresistance in gastric cancer cell lines.Citation19 Splicing factor, serine and arginine rich splicing factor 4 (SRSF4) has been shown to play a role in cisplatin induced pre-mRNA splicing and apoptosis of gastric cancer cells.Citation21
Validation of tubulin beta 3 class III (TUBB3) expression in cisplatin treated HNSCC cells
To determine the role of proteins that were altered upon cisplatin treatment (24 h) in platinum resistance, we compared data from this study with proteins previously linked to platinum resistance. A study by Huang D., et al., has reported over 900 proteins to be associated with platinum resistance.Citation22 Of the 343 proteins that were differentially regulated upon cisplatin treatment in this study, we observed an overlap of over 45 proteins with those known to be associated with platinum resistance (). This serves as a proof-of-concept that these proteins undergo alterations as early as 24 h and could play a role in developing platinum resistance. Tubulin beta 3 class III (TUBB3) is one of the proteins that was found to be common and is also reported in literature to be overexpressed upon acquisition of platinum resistance.Citation23–25 Mass spectrometry analysis revealed upregulation of TUBB3 in response to cisplatin treatment (24 h) compared to control cell lines. We further validated the expression of TUBB3 in CAL27 and Det562 cells treated with cisplatin for 24 h and in cells that have acquired resistance to cisplatin on chronic treatment.
Figure 4. (a) Venn diagram showing the list of proteins that were common between this study and proteins associated with platinum resistance. (b-c) Development of cisplatin resistant cell lines. CAL27 and Det562 cells were treated with cisplatin at their IC50 for 3 months. The IC50 of both the cells were found to increase with chronic cisplatin treatment indicating acquisition of resistant phenotype. (d) CAL27 and Det562 cisplatin resistant cells exhibited higher cellular proliferation rate compared to untreated or parental cell lines; ((e-f)) Expression of TUBB3 in untreated and cisplatin treated CAL27 and Det562. TUBB3 was found to be overexpressed in cells exposed to cisplatin compared to untreated cells. Cisplatin resistant cells (R) also showed a higher expression of TUBB3 compared to cells treated with cisplatin for 24 h. The expression of TUBB3 was found to be statistically significant in Det562 cells. β- actin was used as loading control.
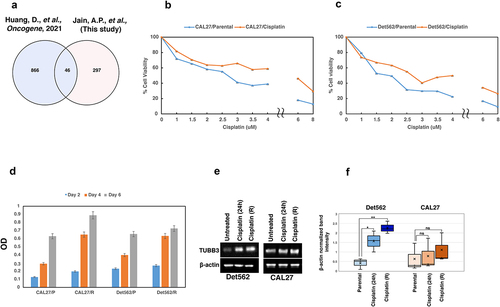
Cisplatin resistant, CAL27 and Det562 cell lines were developed as described previously.Citation26 Briefly, CAL27 and Det562 cells were treated with cisplatin at their IC50 for 72 h. Following this, the cells were maintained in cisplatin-free media until the surviving cells recovered. This process was repeated for 3 months. The IC50 of parental and cisplatin treated cells was then determined by MTT assay. As shown in , chronic treatment of CAL27 and Det562 cells with cisplatin led to an increase in IC50 compared to the parental cells indicating the acquisition of cisplatin resistance phenotype. Further, we observed that cisplatin resistant, CAL27 and Det562 cells exhibited higher cellular proliferation rate compared to untreated or parental cells (). Western blot analysis confirmed the overexpression of TUBB3 in cells treated with cisplatin for 24 h, as well as in cisplatin resistant CAL27 and Det562 cell lines compared to untreated cells (). These results indicate that subsets of proteins implicated in protection against cisplatin are activated as early as 24 h and these could be molecules that enable cells to evade cisplatin induced damage.
Discussion
Studies have shown that determining response of cells to short-term drug treatment will provide in-sights into the i) mechanisms of action of drugs and ii) adaptive mechanisms employed by cells to counteract the anti-proliferative effects of drugs.Citation4,Citation6 In the past, mass spectrometry-based proteomic approaches have been successfully employed to elucidate the mechanisms of action of various drugs.Citation4,Citation6,Citation7 However, the precise mechanisms of action of cisplatin are not completely understood, despite its widespread use in cancer treatment. The current study demonstrates the successful application of label-free quantitative proteomic approach to examine the alterations in the cellular proteome of multiple HNSCC cell lines induced by short-term cisplatin treatment. Using more than one cell type in a study will lead to ii) high proteome coverage, ii) an in-depth knowledge on the signaling mechanisms induced by cisplatin, and iii) enhanced statistical power.
Exposure of HNSCC cell lines to cisplatin for 24 h resulted in the alterations of various cellular processes including DNA damage response pathway, translation and mRNA splicing. The response of cells to a genotoxic insult can result either in apoptosis of cells due to damaged DNA or repair of DNA damage that enables cell survival. A literature-based analysis of data from this study revealed several proteins implicated in DNA damage response and repair to be altered. Proteins such as RAD50 double strand break repair protein (RAD50), Aldo-keto reductase family 1 member B10 (AKR1B10), aldehyde dehydrogenase 2 family member (ALDH2), and S100 calcium binding protein A8 (S100A8) implicated in DNA damage response pathway were found to be altered within 24 h of treatment. Further, we observed proteins implicated in translation and mRNA splicing that are associated with cisplatin resistance to be enriched. These results indicate that the cells activate protective mechanisms to cisplatin as early as 24 h of exposure and these proteins may contribute to the development of cisplatin resistance.
In addition to proteins implicated in regulating diverse cellular functions, a subset of differentially expressed proteins from our study was found to be associated with platinum resistance. LC-MS/MS analysis revealed several microtubule proteins such as TUBB3, TUBB4A, and TUBB8 to be altered upon cisplatin treatment in HNSCC cell lines. Microtubules are the cytoskeletal proteins implicated in various processes including cell motility, intracellular transport, and mitosis.Citation27 TUBB3 is reported to be expressed in neuronal cells, melanocytes, and follicular lymphoid cells.Citation28 In cancers such as HNSCC, lung, ovarian, and bladder cancer, TUBB3 is shown to be overexpressed.Citation29–32 Patients with high expression of TUBB3 exhibit poor survival rate and resistance to docetaxel, paclitaxel- and cisplatin-based chemotherapies.Citation24,Citation33–37 Head and neck cancer patients positive for TUBB3 expression and undergoing induction chemotherapy with 5-Flurouracil (5-FU) and cisplatin and 5-FU, cisplatin and docetaxel exhibited lower response rate. In addition, they also showed a higher disease progression and reduction in overall survival rate compared to TUBB3-negative patients.Citation32 These studies indicate the significance of TUBB3 as a predictive and prognostic marker not only in HNSCC patients but also in other cancers. In the current study, we observed upregulation of TUBB3 upon cisplatin treatment. Western blot analysis further confirmed the overexpression of TUBB3 upon short-term and chronic exposure to cisplatin in CAL27 and Det562 cell lines. The exact mechanisms of role of TUBB3 in inducing cisplatin resistance is not known. However, a study by Gan, P.P., et al., has shown that silencing of TUBB3 sensitizes non-small cell lung carcinoma (NSCLC) cells to DNA damaging agents including cisplatin and induces apoptosis in response to cisplatin treatment indicating its role in drug sensitivity.Citation38 Though the current study provides a clue that TUBB3 can serve as a reliable predictive marker of cisplatin sensitivity in HNSCC patients, additional in vitro and in vivo studies are warranted to ascertain its clinical utility. Targeted mass spectrometry-based validation of TUBB3 in large cohort of samples from HNSCC patients undergoing neoadjuvant therapy and from chemotherapy responders and non-responders will enhance the knowledge on utility of TUBB3 as a predictive marker. In addition to TUBB3, the present study, however, does not rule out the role of other molecules in inducing cisplatin resistance.
Conclusions
In summary, this study reveals the molecular level alterations that occur in response to short-term treatment to cisplatin in head and neck cancer. We demonstrate that certain molecules that play a role in the development of cisplatin resistance are activated as early as 24 h. These could be the molecules that enable cells to adapt to cisplatin and escape cell death resulting in the acquisition of resistance phenotype. However, more in vivo and in vitro studies are warranted to understand the role of these proteins in inducing cisplatin resistance and their role as potential predictive markers of cisplatin therapy.
Authors’ contributions
A.P.J and V.N conceived and designed the study. A.P.J. and V.N. were involved in cell culture and performed all assays. V.G. F.A.B and G.D. performed proteomics experiments, fractionation and carried out mass spectrometric analysis of samples. S.M. carried validation experiments. A.P.J., G.D., and V.N. prepared the manuscript, figures and tables. All the authors have read and approved the final manuscript.
Supplemental Material
Download MS Excel (3.3 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplemental material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/23723556.2024.2328873
Additional information
Funding
References
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–9. doi:10.1016/j.ejphar.2014.07.025.
- Pietras P, Aulas A, Fay MM, Leśniczak-Staszak M, Sowiński M, Lyons SM, Szaflarski W, Ivanov P. Translation inhibition and suppression of stress granules formation by cisplatin. Biomed Pharmacother. 2022;145:112382. doi:10.1016/j.biopha.2021.112382.
- Bostan M, Petrică‑Matei GG, Ion G, Radu N, Mihăilă M, Hainăroşie R, Braşoveanu L, Roman V, Constantin C, Neagu M. et al. Cisplatin effect on head and neck squamous cell carcinoma cells is modulated by ERK1/2 protein kinases. Exp Ther Med. 2019;18(6):5041–5051. doi:10.3892/etm.2019.8139.
- Ebhardt HA, Root A, Liu Y, Gauthier NP, Sander C, Aebersold R. Systems pharmacology using mass spectrometry identifies critical response nodes in prostate cancer. NPJ Syst Biol Appl. 2018;4(1):26. doi:10.1038/s41540-018-0064-1.
- Musiani D, Hammond DE, Cirillo L, Erriquez J, Olivero M, Clague MJ, Di Renzo MF. PIM2 kinase is induced by cisplatin in ovarian cancer cells and limits drug efficacy. J Proteome Res. 2014;13(11):4970–4982. doi:10.1021/pr500651n.
- Mitchell DC, Kuljanin M, Li J, Van Vranken JG, Bulloch N, Schweppe DK, Huttlin EL, Gygi SP. A proteome-wide atlas of drug mechanism of action. Nat Biotechnol. 2023;41(6):845–857. doi:10.1038/s41587-022-01539-0.
- Frejno M, Meng C, Ruprecht B, Oellerich T, Scheich S, Kleigrewe K, Drecoll E, Samaras P, Hogrebe A, Helm D. et al. Proteome activity landscapes of tumor cell lines determine drug responses. Nat Commun. 2020;11(1):3639. doi:10.1038/s41467-020-17336-9.
- Frejno M, Zenezini Chiozzi R, Wilhelm M, Koch H, Zheng R, Klaeger S, Ruprecht B, Meng C, Kramer K, Jarzab A. et al. Pharmacoproteomic characterisation of human colon and rectal cancer. Mol Syst Biol. 2017;13(11):951. doi:10.15252/msb.20177701.
- Borse V, Konwar AN, Buragohain P. Oral cancer diagnosis and perspectives in India. Sens Int. 2020;1:100046. doi:10.1016/j.sintl.2020.100046.
- Griso AB, Acero-Riaguas L, Castelo B, Cebrian-Carretero JL, Sastre-Perona A. Mechanisms of cisplatin resistance in HPV negative head and neck squamous cell carcinomas. Cells. 2022;11(3):561. doi:10.3390/cells11030561.
- Nanjappa V, Renuse S, Sathe GJ, Raja R, Syed N, Radhakrishnan A, Subbannayya T, Patil A, Marimuthu A, Sahasrabuddhe NA. et al. Chronic exposure to chewing tobacco selects for overexpression of stearoyl-CoA desaturase in normal oral keratinocytes. Cancer Biol Ther. 2015;16(11):1593–1603. doi:10.1080/15384047.2015.1078022.
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu D, Prakash A, Frericks-Zipper A, Eisenacher M. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50(D1):D543–D552. doi:10.1093/nar/gkab1038.
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi:10.1093/bioinformatics/btp101.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303.
- Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, Evelo CT. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol. 2015;11(2):e1004085. doi:10.1371/journal.pcbi.1004085.
- Liu YQ, Zhang GA, Zhang BC, Wang Y, Liu Z, Jiao Y-L, Liu N, Zhao Y-R. Short low concentration cisplatin treatment leads to an epithelial mesenchymal transition-like response in DU145 prostate cancer cells. Asian Pac J Cancer Prev. 2015;16(3):1025–1028. doi:10.7314/APJCP.2015.16.3.1025.
- Zou J, Zhu L, Jiang X, Wang Y, Wang X, Chen B. Curcumin increases breast cancer cell sensitivity to cisplatin by decreasing FEN1 expression. Oncotarget. 2018;9(13):11268–11278. doi:10.18632/oncotarget.24109.
- Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen. 2017;58(5):235–263. doi:10.1002/em.22087.
- Huang H, Han Y, Zhang C, Wu J, Feng J, Qu L, Shou C. HNRNPC as a candidate biomarker for chemoresistance in gastric cancer. Tumour Biol. 2016;37(3):3527–3534. doi:10.1007/s13277-015-4144-1.
- Taoka Y, Matsumoto K, Ohashi K, Minamida S, Hagiwara M, Nagi S, Saito T, Kodera Y, Iwamura M. Protein expression profile related to cisplatin resistance in bladder cancer cell lines detected by two-dimensional gel electrophoresis. Biomed Res. 2015;36(4):253–261. doi:10.2220/biomedres.36.253.
- Gabriel M, Delforge Y, Deward A, Habraken Y, Hennuy B, Piette J, Klinck R, Chabot B, Colige A, Lambert C. et al. Role of the splicing factor SRSF4 in cisplatin-induced modifications of pre-mRNA splicing and apoptosis. BMC Cancer. 2015;15(1):227. doi:10.1186/s12885-015-1259-0.
- Huang D, Savage SR, Calinawan AP, Lin C, Zhang B, Wang P, Starr TK, Birrer MJ, Paulovich AG. A highly annotated database of genes associated with platinum resistance in cancer. Oncogene. 2021;40(46):6395–6405. doi:10.1038/s41388-021-02055-2.
- Kato A, Naitoh I, Naiki-Ito A, Hayashi K, Okumura F, Fujita Y, Sano H, Nishi Y, Miyabe K, Inoue T. et al. Class III β-tubulin expression is of value in selecting nab-paclitaxel and Gemcitabine as first-line therapy in unresectable pancreatic cancer. Pancreas. 2022;51(4):372–379. doi:10.1097/MPA.0000000000002032.
- Maahs L, Sanchez BE, Gupta N, Van Harn M, Barrack ER, Reddy P-V, Hwang C. Class III β-tubulin expression as a predictor of docetaxel-resistance in metastatic castration-resistant prostate cancer. PLoS One. 2019;14(10):e0222510. doi:10.1371/journal.pone.0222510.
- Miyata Y, Matsuo T, Nakamura Y, Yasuda T, Ohba K, Takehara K, Sakai H. Expression of class III beta-tubulin predicts prognosis in patients with cisplatin-resistant bladder cancer receiving paclitaxel-based second-line chemotherapy. Anticancer Res. 2018;38(3):1629–1635.
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546(7658):431–435. doi:10.1038/nature22794.
- Valiron O, Caudron N, Job D. Microtubule dynamics. Cell Mol Life Sci. 2001;58(14):2069–2084. doi:10.1007/PL00000837.
- Person F, Wilczak W, Hube-Magg C, Burdelski C, Möller-Koop C, Simon R, Noriega M, Sauter G, Steurer S, Burdak-Rothkamm S. et al. Prevalence of βIII-tubulin (TUBB3) expression in human normal tissues and cancers. Tumour Biol. 2017;39(10):1010428317712166. doi:10.1177/1010428317712166.
- Choi JW, Kim Y, Lee JH, Kim YS. Expression of beta-tubulin isotypes in urothelial carcinoma of the bladder. World J Urol. 2014;32(2):347–352. doi:10.1007/s00345-012-0993-z.
- Ferrandina G, Zannoni GF, Martinelli E, Paglia A, Gallotta V, Mozzetti S, Scambia G, Ferlini C. Class III β-tubulin overexpression is a marker of poor clinical outcome in advanced ovarian cancer patients. Clin Cancer Res. 2006;12(9):2774–2779. doi:10.1158/1078-0432.CCR-05-2715.
- Koh Y, Jang B, Han SW, Kim T-M, Oh D-Y, Lee S-H, Kang CH, Kim D-W, Im S-A, Chung DH. et al. Expression of class III beta-tubulin correlates with unfavorable survival outcome in patients with resected non-small cell lung cancer. J Thorac Oncol. 2010;5(3):320–325. doi:10.1097/JTO.0b013e3181ce684f.
- Koh Y, Kim TM, Jeon YK, Kwon T-K, Hah JH, Lee S-H, Kim D-W, Wu H-G, Rhee C-S, Sung M-W. et al. Class III β-tubulin, but not ERCC1, is a strong predictive and prognostic marker in locally advanced head and neck squamous cell carcinoma. Ann Oncol. 2009;20(8):1414–1419. doi:10.1093/annonc/mdp002.
- Di Bartolomeo M, Raimondi A, Cecchi F, Catenacci DVT, Schwartz S, Sellappan S, Tian Y, Miceli R, Pellegrinelli A, Giommoni E. et al. Association of high TUBB3 with resistance to adjuvant docetaxel-based chemotherapy in gastric cancer: translational study of ITACA-S. Tumori. 2021;107(2):150–159. doi:10.1177/0300891620930803.
- Na HY, Park M, Kim YA, Won JK, Park YJ, Shin SA, Lee S, Oh S, Kim JE. Expression of class III beta-tubulin is associated with invasive potential and poor prognosis in thyroid carcinoma. J Clin Med. 2020;9(12):3830. doi:10.3390/jcm9123830.
- Sekino Y, Han X, Babasaki T, Miyamoto S, Kitano H, Kobayashi G, Goto K, Inoue S, Hayashi T, Teishima J. et al. TUBB3 is associated with high-grade histology, poor prognosis, p53 expression, and cancer stem cell markers in clear cell renal cell carcinoma. Oncology. 2020;98(10):689–698. doi:10.1159/000506775.
- Kamath K, Wilson L, Cabral F, Jordan MA. BetaIII-tubulin induces paclitaxel resistance in association with reduced effects on microtubule dynamic instability. J Biol Chem. 2005;280(13):12902–12907. doi:10.1074/jbc.M414477200.
- Stengel C, Newman SP, Leese MP, Potter BV, Reed MJ, Purohit A. Class III beta-tubulin expression and in vitro resistance to microtubule targeting agents. Br J Cancer. 2010;102(2):316–324. doi:10.1038/sj.bjc.6605489.
- Gan PP, Pasquier E, Kavallaris M. Class III beta-tubulin mediates sensitivity to chemotherapeutic drugs in non small cell lung cancer. Cancer Res. 2007;67(19):9356–9363. doi:10.1158/0008-5472.CAN-07-0509.

