ABSTRACT
ATG1/ATG13, a core kinase complex regulator in the macroautophagy/autophagy machinery during autophagosome formation, is modulated in a sophisticated manner by posttranslational ubiquitination to determine proper autophagy levels in eukaryotic cells. However, the mechanisms that regulate the stability and activity of this complex remain elusive. We recently identified two negative regulators of autophagy, 14-3-3λ and 14-3-3k, that help redundantly regulate autophagy by directly associating with SINAT and ATG13s. The specific interaction between the molecular adaptors 14-3-3λ and 14-3-3κ and phosphorylated ATG13a is crucial for SINAT-mediated ubiquitination and degradation of ATG13a, and for maintenance of the ATG1-ATG13 complex. Consistent with the function of 14-3-3s in autophagy, the 14-3-3λ 14-3-3κ double mutant exhibits enhanced tolerance to nutrient deprivation with constitutive induction of autophagy. These findings demonstrate that 14-3-3λ and 14-3-3k coordinate with SINATs to regulate both the homeostasis of ATG13 phosphorylation and the induction of autophagy in plants.
In eukaryotes, autophagy is a conserved process that degrades and recycles unwanted cytoplasmic constituents and damaged organelles. This process occurs in the vacuole or lysosome and is essential for nutrient recycling and cellular housekeeping. As part of the autophagic machinery in plants, a group of four core protein complexes assembled by numerous autophagy-related (ATG) proteins have been identified. Of these, the ATG1-ATG13 protein kinase complex is the most upstream positive regulator of autophagy and is tightly modulated by posttranslational modifications, such as phosphorylation and ubiquitination, during autophagy initiation. However, the underlying molecular mechanism through which the interplay between phosphorylation and ubiquitination regulates plant autophagy remains poorly understood.
In this study [Citation1], we discovered that the highly conserved eukaryotic scaffolding molecules 14-3-3λ and 14-3-3κ specifically associate with phosphorylated ATG13a to modulate SINAT-mediated proteolysis and regulate the interaction between ATG13 and ATG1 in Arabidopsis thaliana, based on the following observations: 1) the molecular adaptors 14-3-3λ and 14-3-3κ physically interact with the E3 ubiquitin ligase SINAT1; 2) plants lacking 14-3-3λ and 14-3-3κ display enhanced tolerance to nutrient deprivation, delayed leaf senescence, and increased starvation-induced autophagic vesicle formation; 3) 14-3-3λ and 14-3-3κ associate with ATG1-ATG13 directly in vitro and in vivo to enhance the association of SINATs with ATG13a and thus modulate SINAT-mediated ubiquitination and degradation of ATG13a in plants; 4) 14-3-3λ and 14-3-3κ specifically associate with the phosphorylated form of ATG13 in vitro and in vivo to modulate its homeostasis and the formation of the ATG1-ATG13 complex; 5) Mutation of 18 putative phosphorylation sites on ATG13a revealed phosphorylation is crucial for the autophagy-mediated tolerance of nutritional deprivation in Arabidopsis. Taken together, these observations suggest that the molecular adaptors 14-3-3λ and 14-3-3κ are involved in the regulation of autophagy dynamics by modulating ATG13 stability and thus the formation of the ATG1-ATG13 kinase complex in plants.
YWHA/14-3-3 proteins are highly conserved scaffold proteins that form homo- and hetero-dimers and bind to specific phosphothreonine and phosphoserine motifs on their targets. Through these interactions, YWHA/14-3-3 proteins are involved in diverse signal transduction events in eukaryotic organisms. In mammals, YWHA/14-3-3 proteins play important roles in autophagy by binding to ATG proteins such as BECN1, PIK3C3/VPS34, ATG9A, and ULK1 to modulate their activity, stability, and regulate their recruitment to autophagosomes. We further found that 14-3-3λ and 14-3-3κ act as negative regulators of autophagy in plants by associating with phosphorylated ATG13 to modulate its proteolysis and regulate the formation of the ATG1-ATG13 complex. This confirms that although the mechanisms of action may vary the regulation of YWHA/14-3-3 protein-mediated autophagy is evolutionarily conserved between mammals and plants.
Our current working model () highlights the function of 14-3-3λ and 14-3-3κ as molecular switches regulating the homeostasis of phosphorylated ATG13 levels and the formation of the ATG1-ATG13 complex under different nutrient conditions. Under nutrient-rich conditions, 14-3-3λ and 14-3-3κ function as adaptors to specifically associate with phosphorylated ATG13 and recruit the E3 ubiquitin ligases SINAT1 and SINAT2 to form a plant-specific 14-3-3-SINAT-ATG1-ATG13 complex. This complex promotes 26S proteasome-mediated ubiquitination and degradation of phosphorylated ATG13. In parallel, the interaction of ATG13a with 14-3-3 proteins decreases its affinity for ATG1 in plants. These two pathways work cooperatively to quickly reduce the overall amount of phosphorylated ATG13 and slow formation of the ATG1-ATG13 complex, thus repressing plant autophagy. This model describes a possible new mechanism for the regulation of autophagy in plants.
Figure 1. A proposed working model of 14-3-3 proteins in plant autophagy. Under nutrient-rich conditions, 14-3-3 proteins inhibit autophagy by recruiting E3 ubiquitin ligases SINAT1 and SINAT2 to degrade phosphorylated ATG13 via the 26S proteasome pathway and also promote the dissociation of the ATG1-ATG13 complex, thus leading to the suppression of autophagy. Under nutrient-deprivation conditions, the dephosphorylation of ATG13a is mediated by TOPP, leading to reduced affinity with 14-3-3 proteins and increased binding with hyperphosphorylated ATG1a, thus facilitating autophagy initiation events in plants.
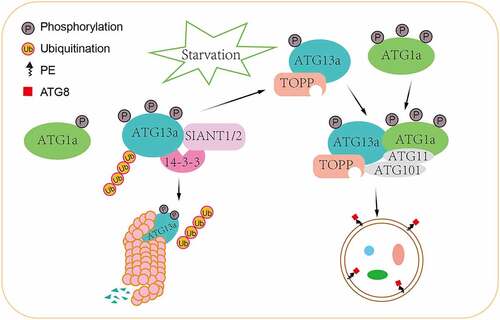
In response to nutrient availability, the phosphorylation status of ATG13 is controlled by the upstream negative regulator TOR/mTOR in yeast and mammals; however, such a regulatory mechanism is not well defined in plants. A previous study showed that the RAPTOR interacts with ATG13a in plants to regulate autophagy, providing evidence supporting a role of TOR in the regulation of plant autophagy. Such evolutionary conservation of TOR-mediated autophagy regulation between mammals and plants suggests that ATG13 may be a substrate of TOR. In this study, we identified 18 phosphorylation sites in ATG13a which may mediate the interaction with TOR under nutrient-rich conditions in plants. Further studies are needed to identify additional upstream regulators as well as the molecular mechanism underlying modulation of ATG13 phosphorylation. Potential upstream regulators of ATG13 in plants include MPK3 and MPK6.
Under nutrient deprivation conditions, ATG13a is directly dephosphorylated by TOPPs, leading to reduced affinity with 14-3-3 proteins, and increased interaction with hyperphosphorylated ATG1a to initiate autophagy (). Our previous study showed that a positive kinase regulator, SnRK1, may facilitate autophagy initiation events by phosphorylating ATG1 in plants. Although the direct interaction of ATG1-ATG13 and SnRK1 was not detected in plants, increased ATG1 phosphorylation is associated with overexpression of the α-subunit of SnRK1, KIN10, indicating that SnRK1 is involved in phosphorylation of the ATG1-ATG13 complex in plants. Further investigation of how the energy sensors TOR and SnRKs initiate autophagy will allow us to obtain a deeper understanding of the mechanisms by which energy signaling regulates autophagy in plants.
The scaffold proteins TRAF1a and TRAF1b can act as positive or negative regulators to modulate autophagy dynamics by forming plant-type TRAFsome complexes with ATG13 or ATG6 and SINATs. These complexes modulate the ubiquitination and degradation of ATG13 and ATG6 under different nutrient conditions, indicating the crosstalk between 14-3-3s and TRAF1s in modulating plant autophagy. Further investigation of the molecular mechanism by which these two scaffold proteins regulate autophagy dynamics in plants is required. Similar to ATG13 and ATG6, ATG1 also undergoes 26S proteasome-dependent degradation under nutrient-deprivation conditions; the regulation mechanism that SINATs or other E3 Ub ligases induce ATG1 degradation also remains to be uncovered.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Key Realm R&D Program of Guangdong Province (Project 2020B0202090001; 2020B0202080001), the Natural Science Foundation of Guangdong Province (Project 2022A1515012500; 2018A030313210), and the Laboratory of Lingnan Modern Agriculture (Project NG2021002; NT2021010).
Reference
- Qi H, Lei X, Wang Y, Yu S, Liu T, Zhou SK, Chen JY, Chen QF, Qiu RL, Jiang L, and Xiao S. 14-3-3 proteins contribute to autophagy by modulating SINAT-mediated degradation of ATG13. Plant Cell. 2022. 34: 4857–5.

