Degradation of proteins in cells is carried out by two major systems: the ubiquitin proteasome system (UPS) and autophagy. Macroautophagy is a type of autophagy in which vesicles in the cytoplasm, called autophagosomes, engulf and degrade long-lived cytosolic proteins and damaged organelles to harvest the building blocks for cell survival. Although macroautophagy (referred to as autophagy hereon) is classically activated by nutrient deprivation, the unfolded protein response (UPR) is another factor that can lead to its induction. IRE1/ERN1 (inositol-requiring enzyme 1), a serine/threonine kinase and an endoribonuclease, is one of the three ER transmembrane proteins that detect the accumulation of misfolded proteins in this organelle and activate UPR in mammalian cells while it is the only one present in yeast. IRE1 can activate autophagy by inducing the JNK (c-Jun N-terminal kinase) signalling pathway, which promotes degradation of the mammalian/mechanistic target of rapamycin complex 1 (mTORC1) complex, a negative autophagy regulator. This leads to the clearance of misfolded or aggregated proteins in the ER and elsewhere. In case of proteasomal degradation, IRE1 activates the XBP1s signaling pathway, which in turn promotes retrotranslocation from the ER and degradation of misfolded or aggregated proteins by the cytoplasmic proteasome. Downregulation of the IRE1α-XBP1 axis in Huntington’s disease (HD) and amyotrophic lateral sclerosis inhibits autophagy, accelerating disease progression in mice models. In specific proteinopathy models such as those characterizing Huntington’s disease, abnormal ER stress can also block autophagy. Under chronic ER stress caused by the accumulation of aggregated proteins, the capacity of the ER quality control system is exceeded. This is accompanied by a lowering in the activation of the UPR pathways. What is triggering this dampening in the UPR is still unknown. Prolonged ER stress leads to the inhibition of the autophagic flux via IRE1-TRAF2 (tumour necrosis factor receptor-associated factor 2) pathway and impairs the autophagic clearance of mutant huntingtin aggregates in neuronal cells. The ability of IRE1 to regulate both autophagy and proteasomal degradation suggests that it may be a promising target for the development of therapies that alleviate proteotoxic stress and promote protein homeostasis in pathological situations. Autophagy is known to be involved in the degradation of mutant huntingtin. Activation of autophagy through pharmacological interventions has demonstrated beneficial effects in HD models.
Our recent study [Citation1] was initiated with two goals; (i) to identify whether IRE1 has a direct role in the regulation of autophagy under conditions of proteotoxic stress, and (ii) to identify a possible regulator of dampened stress response during unmitigated or prolonged stress. In particular, we monitored the role of Ire1 in a humanized yeast model of HD. Prolonged aggregation of mutant huntingtin fragment (NT-103Q) fused with EGFP was monitored by inducing the expression of the aggregation-prone protein in yeast with 2% galactose for up to 20 h. Aggregation of NT-103Q was monitored using a specific polyglutamine antibody. The effect of Ire1 was monitored by either deleting the encoding gene or inactivation of its enzymatic activity using two structurally distinct classes of inhibitors, 4μ8C and folinic acid. The addition of these inhibitors resulted in a pattern of protein aggregation similar to that observed in ire1Δ cells. Protein aggregation assessed by filter retardation assay was seen to decrease at longer time points (16–20 h) upon deletion/inactivation of Ire1 in comparison to shorter periods (8 h). Treatment with the anionic detergent sodium dodecyl sulphate (SDS), to discriminate between amorphous detergent-soluble from fibrillar detergent-insoluble aggregates, revealed that at 20 h, the level of fibrillar aggregates was lower in cells lacking Ire1 signalling, suggesting a decline in fibrillar aggregate formation in the absence of Ire1. Reduction of protein aggregation can arise due to lower protein synthesis or its faster degradation. Synthesis of wild type huntingtin fragment harbouring a polyQ stretch of 25 residues was similar in wild type and ire1Δ cells, suggesting that attenuated protein expression was not the cause of accelerated degradation. In contrast, the mutant NT-103Q was found to be degraded faster in ire1Δ cells upon switching off of protein expression by removal of galactose, suggesting that the protein degradation mechanism(s) was inordinately activated in these cells.
To investigate the reason for stronger decline in protein degradation at longer time points in Ire1-depleted cells, we monitored the three major stress response pathways: UPR, UPS and autophagy. Higher aggregation, which is seen in wild-type cells, did not lead to a stronger induction of the UPR. Although splicing of Hac1 (Homologous to Atf/Creb1), a client of Ire1 and a marker of UPR, was significantly increased upon protein aggregation, suggesting the activation of ER stress response, the splicing activity also declined upon prolonged aggregation indicating that activation of UPR was mitigated in this situation. As expected, no Hac1 splicing was observed in ire1Δ cells. The expression of Rpn4 (Regulatory Particle Non-ATPase), encoding a transcription factor regulating proteasome biogenesis, was upregulated upon aggregation in both wild-type and ire1Δ cells although the upregulation was lower in wild-type cells at later time points. Prolonged aggregation led to increased proteasomal activity (chymotrypsin-like activity) in ire1Δ cells in comparison to wild-type cells. Cytoplasmic and ER-specific degron sequences were also used to monitor proteasomal function. High proteasomal activity was detected with both sets of reporters in Ire1-inhibited cells. Notably, the proteasomal activity decreased significantly at 20 h post-induction in wild-type cells when the cytoplasmic degron reporter construct was employed. Thus, the enhanced proteasomal activity in ire1Δ cells, possibly via ER-associated degradation (ERAD), correlated with decreased protein aggregation, suggesting a potential role for the proteasome in reducing aggregation.
Prolonged ER stress is an autophagy inducer. Thus, we explored the involvement of Ire1 in autophagy in response to prolonged aggregation. This is a major pathway by which mutant huntingtin is reported to be degraded in cells. Atg8 is a core autophagy-related (Atg) protein that is upregulated upon autophagy induction. Downregulation of ATtg8 expression was seen in wild-type cells while no such reduction was observed upon deletion of Ire1. Interestingly and in agreement with previous reports, treatment with 1,4-dithiothreitol (DTT) led to significant upregulation of Atg8 in both wild-type and ire1Δ cells. This suggests that while the presence of Ire1 dampens the activation of autophagy upon prolonged stress, the signaling pathways triggering autophagy upon nutrient starvation or UPR are distinct. Autophagic activity was reduced in both wild-type and ire1Δ cells at longer time points of aggregation. However, the decline was steeper in wild-type cells. Autophagy was monitored by GFP-Atg8 processing assay. The amount of total GFP (cleaved and tagged) was higher in ire1Δ cells and matched the upregulation pattern of Atg8 in these cells. Autophagic flux was determined by measurement of accumulation of free GFP following autophagic degradation of GFP-Atg8. No change was observed in wild-type cells while ire1Δ cells exhibited higher induction of autophagy at longer time points, indicating the compensatory role of autophagy in protein clearance. Importantly, higher autophagic activity in ire1Δ cells correlated well with lower levels of protein aggregates at longer time points. These findings suggest that Ire1 plays a critical role in modulating dynamics of aggregate turnover of mutant huntingtin.
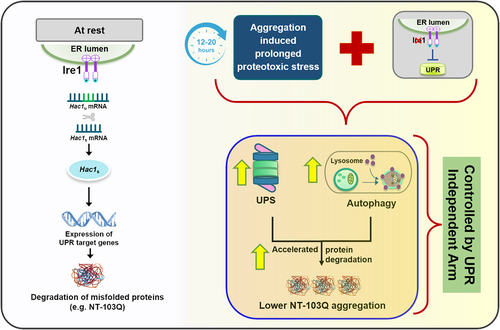
Several studies have shown conclusively that exposure to unmitigated stress does not lead to an infinite activation of stress responses in cells. Whether this is due to a failure of the cellular protein quality control becoming overwhelmed or to a programmed response, has not been uncovered yet. As shown in , our study suggests that Ire1, sitting at the crossroad of UPR, UPS and autophagy, may be a key component of the complex network required to maintain the critical balance between stress and the cell’s adaptive response to it.
Figure 1. Ire1 acts as a lid to regulate proteostasis response during chronic stress in yeast cells. In the resting stage, Ire1 activates the UPR upon accumulation of misfolded mutant huntingtin (NT-103Q). We show that in the absence of Ire1, compensatory mechanisms such as autophagy and proteasomal function are upregulated upon prolonged stress and reduce aggregation of NT-103Q.
List of abbreviations
| HD | = | Huntington’s disease |
| Ire1 | = | Inositol-requiring enzyme 1 |
| NT-103Q | = | N-terminal huntingtin carrying 103 glutamine residues |
| UPS | = | Ubiquitin proteasome system |
| UPR | = | Unfolded protein response |
Additional information
Funding
Reference
- Das, E., K.K. Sahu, and I. Roy, The functional role of Ire1 in regulating autophagy and proteasomal degradation under prolonged proteotoxic stress, FEBS J, 2023, 290, 3270–5.

