ABSTRACT
Macroautophagy/autophagy delivers cytoplasmic constituents to the lysosome/vacuole for degradation and recycling, wherein regulates diverse aspects of cellular homeostasis. However, more is needed to know how autophagy activity is fine-tuned at the organismal level to optimize plant fitness in response to developmental and environmental cues. We recently revealed that both basal autophagy and phosphate (Pi) starvation-induced autophagy are required to maintain Pi homeostasis by modulating the expression of PHOSPHATE TRANSPORTER 1 (PHT1) Pi transporters. While Pi limitation preferentially increases the number of autophagic structures and the autophagic flux in the differential zone of the Arabidopsis primary root, Pi starvation-induction of ATG8f and ATG8h expression contributes to the control of autophagic flux in the Pi-deplete root and may participate in promoting root branching for resources exploration.
Abbreviations: ATG: autophagy related; PHT1: PHOSPHATE TRANSPORTER 1; Pi: phosphate
As sessile organisms, plants constantly encounter and cope with changing environmental conditions. Meanwhile, roots perceive and transmit the external stimuli into the reprogramming of gene expression at the cellular level and, as a result, the organization of the root system architecture. For example, under the deprivation of mineral nutrients, shoots allocate more nutrients and biomass as source organs to roots. In contrast, roots are dedicated to foraging the soil for resources to sustain their proliferative capacity as sink organs. In this regard, autophagy, a multistep degradation pathway, mediates the delivery of unneeded cytoplasmic components to the vacuole and is closely associated with nutrient recycling and remobilization under basal and stressed conditions. Although the physio-developmental role of autophagy in roots is less explored, autophagy activity markedly varies according to the differentiation stage and cell type.
With emerging evidence supporting the notion that autophagy has differential effects in source and sink organs, we argued that the interpretations of earlier studies of autophagy without separately considering shoot and root parts might obscure our understanding of the cellular function of autophagy. Consistent with this notion, we recently revealed that phosphate (Pi) shortage does not drastically alter the autophagic flux in Arabidopsis whole roots, as estimated by the proteolytic cleavage of GFP-ATG8f and the vacuolar degradation of all endogenous ATG8 isoforms [Citation1]. By contrast, imaging analyses of autophagosomes or autophagic bodies, traced as GFP-ATG8f-labeled puncta in root epidermal or cortical cells, suggested that the formation of autophagic structures and the autophagic flux are preferentially increased in the differential zone of primary roots following Pi depletion. These findings corroborate the notion that Pi starvation-induced autophagy appears to diverge between different root cell types.
Using the Arabidopsis wild-type (WT) and autophagy-deficient (atg) seedlings grown under Pi-sufficient and Pi-deficient conditions, we showed that basal autophagy and Pi starvation-induced autophagy participate in Pi homeostasis through different mechanisms (). When Pi is replete, atg mutants display smaller meristem sizes, reduced shoot Pi levels and decreased shoot:root ratios in Pi distribution. By contrast, atg mutants unexpectedly show impaired root cell division and expansion and enhanced root Pi levels when Pi is limited. In Arabidopsis, the plasma membrane–localized PHT1 family has nine members, of which PHT1;1 and PHT1;4 play a predominant role in Pi uptake. The disturbed Pi homeostasis in atg mutants can be partially attributed to dysregulated expression of PHOSPHATE TRANSPORTER 1 (PHT1) transporters. We surmised that in the Pi-limited atg mutants, impaired remobilization of Pi might demand more Pi uptake by roots as sinks via a yet-to-be-identified route. Counterintuitively, defective autophagy facilitates, rather than inhibits, the degradation of PHT1 transporters under Pi starvation (). In contrast to basal autophagy, which may surveillance the translation of PHT1 transporters, Pi starvation-induced autophagy enhances a higher expression level of PHT1 transporters, likely via unconventional autophagy pathways (). In mammalian cells, withdrawal of glucose promotes autophagy-dependent and retromer-mediated recycling of glucose transporter to the plasma membrane. Our finding about the interplay between autophagy and the expression of PHT1 transporters opens a new research line to elucidate how autophagy engages in the membrane trafficking of plasma membrane proteins in plants.
Figure 1. Hypothetical models depicting multifaceted roles of autophagy in regulating the expression of PHT1 transporters and ATG8f/h-dependent lateral root formation. (A) Autophagy modulates the expression level of PHT1 transporters. Under Pi sufficiency, autophagy may positively regulate the translation of PHT1 transporters (i). Following Pi deficiency, an increased number of autophagosomes may contribute to the unconventional trafficking of de novo-synthesized PHT1 transporters en route to the plasma membrane, e.g., autophagy-dependent exocytosis (ii) or the trafficking of endocytosed PHT1 transporters back to the plasma membrane through the recycling endosomes (iii), which are otherwise sorted into the late endosomes for vacuolar degradation. (B) Pi starvation-induction of ATG8f and ATG8h is required to maintain the autophagic flux in the Pi-deplete root, which may mediate the lateral root development via hormone signaling-dependent or -independent selective autophagy.
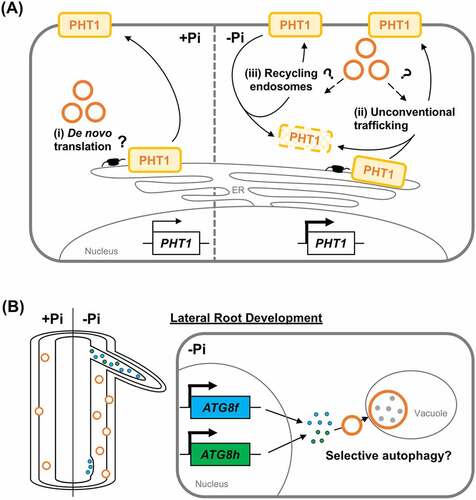
Plants can maximize the root’s surface area for nutrient acquisition by increasing root branching. Phenotypical analyses of atg mutants also suggest that autophagy modulates lateral root formation. Unlike a single ATG8 gene in yeast and algae, the Arabidopsis ATG8 gene family possesses nine genes. We have revealed in a complementary study that A. thaliana ATG8f and ATG8h are progressively upregulated in roots during Pi limitation. Intriguingly, loss of AtATG8f and AtATG8h results in decreased ATG8 proteins associated with membranes[Citation2]. The atg8f/atg8h double mutants also exhibited suppressed root branching and reduced autophagic fluxes in the Pi-depleted roots. These results indicated that these two ATG8 isoforms substantially contribute to the cellular pools of ATG8 proteins in the root and are critical for lateral root formation. As the expression of AtATG8f and AtATG8h mainly overlaps in the primary root stele and is detected at different stages of the lateral root development, we hypothesize that Pi starvation-induction of ATG8f and ATG8h expression fine-tunes root cell type-specific autophagic activity (). It is known that Pi starvation stimulates the formation of lateral roots via crosstalk with hormone signaling. It is, therefore, plausible that selective autophagy-mediated turnover of proteins implicated in hormone signaling dictates lateral root development. Nevertheless, given that the function of ATG8 proteins encompasses selective cargo recruitment, autophagosome–lysosome fusion, and autophagy-independent functions without ATG8 being modified, it remains to unravel how ATG8 isoforms carry out diverse roles on a molecular basis in the root.
Despite our two recent studies unveiling autophagy’s role in maintaining Pi homeostasis and shaping the root system, several questions remain to be addressed. For example, those concern the identity of both the upstream regulators that activate Pi starvation-induced autophagy and the potential cargos subjected to autophagic degradation during Pi starvation in a cell type-specific manner. In unicellular S. cerevisiae and Chlamydomonas, Pi limitation induces autophagy by inhibiting the sensor kinase target of rapamycin (TOR) activity through different genetic components. However, it is unclear whether Pi starvation-induced autophagy in higher plants depends on the TOR signaling, whose sensitivity is also tissue-specific. Moreover, the role of plant hormones in autophagy-mediated modulation of root plasticity awaits additional investigations as well.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chiu CY, Lung HF, Chou WC, et al. Autophagy-Mediated Phosphate Homeostasis in Arabidopsis Involves Modulation of Phosphate Transporters. Plant Cell Physiol. 2023;64(5):519–5. doi: 10.1093/pcp/pcad015
- Lin LY, Chow HX, Chen CH, et al. Role of autophagy-related proteins ATG8f and ATG8h in the maintenance of autophagic activity in Arabidopsis roots under phosphate starvation. Front. Plant Sci. 2023;14. doi: 10.3389/fpls.2023.1018984

