Figures & data
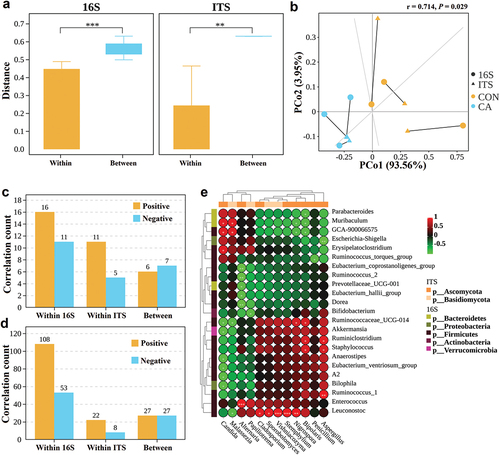
Figure 1. Candida albicans overgrowth induced the disorder of the gut microbiota of bacteria.
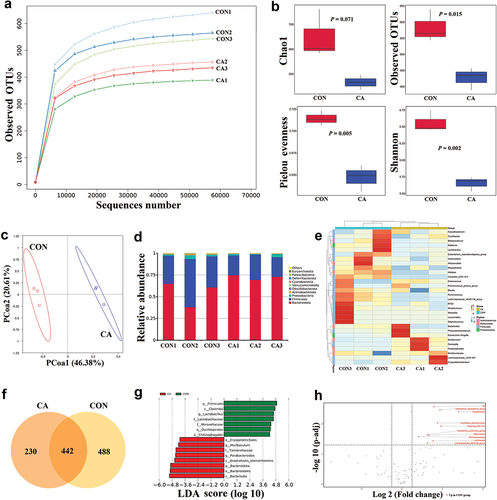
Figure 2. Candida albicans overgrowth induced the disorder of gut microbiota of fungi.
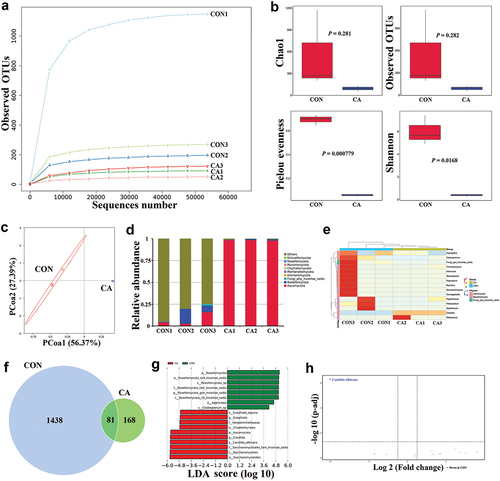
Figure 3. In vivo γδT cell neutralisation partly rescued the bacterial dysbiosis but aggravated the fungal dysbiosis caused by Candida albicans overgrowth.
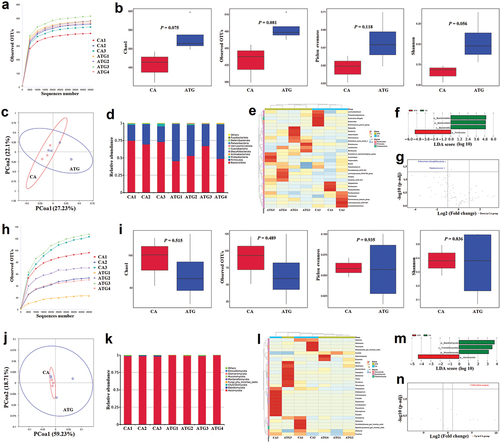
Figure 4. In vivo IL-17A neutralisation aggravated the microbial dysbiosis caused by Candida albicans overgrowth.
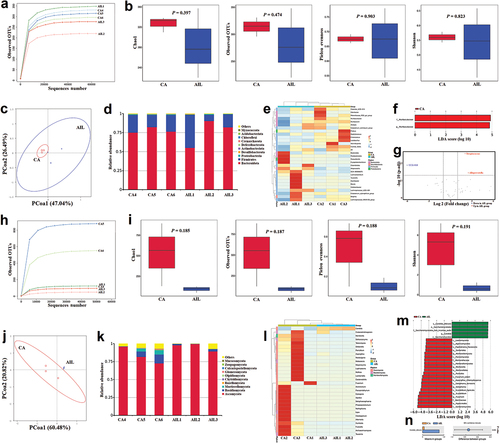
Figure 5. Significant correlations between fungal and bacterial kingdoms were found in the gut microbiome during Candida albicans overgrowth.