Figures & data
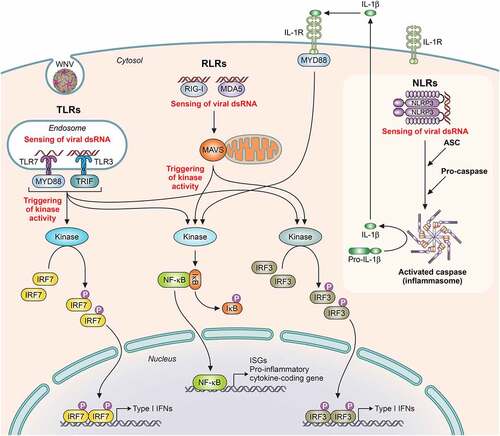
Figure 1. Structure and genome organization of WNV. A) Surface representation of WNV particle displaying the arrangement of E glycoprotein (protein data bank accession 3J0B). B) Schematic view of WNV particle. C) Crystal structure of an E glycoprotein monomer (protein data bank accession 2I69). D) WNV genome organization. The mature proteins produced by cleavage of the single ORF are denoted by boxes. See text for details

Figure 2. Schematic view of WNV infectious cycle. The major steps of WNV infection, including receptor-mediated endocytosis, membrane fusion and genome release, intracellular membrane rearrangements associated with viral replication, immature virion budding into the ER, particle maturation through the secretory pathway, and mature virion release are schematized. See text for details. Electron micrographs to illustrate selected steps of WNV infectious cycle were reproduced under the terms of the creative commons attribution-noncommercial (CC BY-NC 4.0) from [Citation1]
![Figure 2. Schematic view of WNV infectious cycle. The major steps of WNV infection, including receptor-mediated endocytosis, membrane fusion and genome release, intracellular membrane rearrangements associated with viral replication, immature virion budding into the ER, particle maturation through the secretory pathway, and mature virion release are schematized. See text for details. Electron micrographs to illustrate selected steps of WNV infectious cycle were reproduced under the terms of the creative commons attribution-noncommercial (CC BY-NC 4.0) from [Citation1]](/cms/asset/3d5e7c98-b395-4288-9991-3f1ade565dc0/kvir_a_1908740_f0002_oc.jpg)
Figure 3. Phylogenetic analysis based on complete genome nucleotide sequences of the different West Nile virus lineages. GenBank accession numbers, geographic origin and year of isolation of samples are shown. The scale bar depicts genetic distance. The Usutu virus USUV-Spa09 strain was used as out-root

Figure 4. Schematic representation of pathogen recognition receptors (PRRs) activation implicated in WNV infection. Upon binding of viral components, PRRs activation induces a downstream signaling cascade with adaptor proteins implicated and catalytic enzyme activities that promotes the activity of transcription factors, each of which capable of inducing the expression of different target genes, such as Type I IFN genes, ISGs (IFN-stimulated genes) and pro-inflammatory cytokine coding genes. PRRs: TLRs (Toll-like receptors, like TLR-7 and TLR-3), RLR (RIG-I-like receptors, like RIG-I [retinoic acid-inducible gene I protein], and MDA5 [melanoma differentiation antigen 5]) and NLR (NOD-like receptors, like NLRP3). Adaptor proteins: MYD88 (myeloid differentiation 88), TRIF (TIR domain-containing adaptor inducing IFN-β), MAVS (mitochondrial antiviral signaling), and ASC (apoptosis-associated speck-like protein, containing a caspase recruitment domain). Transcription factors: IRF (interferon regulatory factors, like IRF7 and IRF3) and NF-κB (nuclear factor kappa B). Adapted from [Citation265]
![Figure 4. Schematic representation of pathogen recognition receptors (PRRs) activation implicated in WNV infection. Upon binding of viral components, PRRs activation induces a downstream signaling cascade with adaptor proteins implicated and catalytic enzyme activities that promotes the activity of transcription factors, each of which capable of inducing the expression of different target genes, such as Type I IFN genes, ISGs (IFN-stimulated genes) and pro-inflammatory cytokine coding genes. PRRs: TLRs (Toll-like receptors, like TLR-7 and TLR-3), RLR (RIG-I-like receptors, like RIG-I [retinoic acid-inducible gene I protein], and MDA5 [melanoma differentiation antigen 5]) and NLR (NOD-like receptors, like NLRP3). Adaptor proteins: MYD88 (myeloid differentiation 88), TRIF (TIR domain-containing adaptor inducing IFN-β), MAVS (mitochondrial antiviral signaling), and ASC (apoptosis-associated speck-like protein, containing a caspase recruitment domain). Transcription factors: IRF (interferon regulatory factors, like IRF7 and IRF3) and NF-κB (nuclear factor kappa B). Adapted from [Citation265]](/cms/asset/c38b6b84-8dc1-429e-92d2-ad2a81b5160d/kvir_a_1908740_f0004_oc.jpg)
Table 1. Common signs in WNV infected vertebrate species
Table 2. Clinical trials related to passive immunization strategies against WNV
Table 3. Selected clinical trials of WNV vaccines
Table 4. Equine vaccines licensed against WNV
