Figures & data
Figure 1. Establishing a model of ADE in DENV-3 infected THP-1 cells
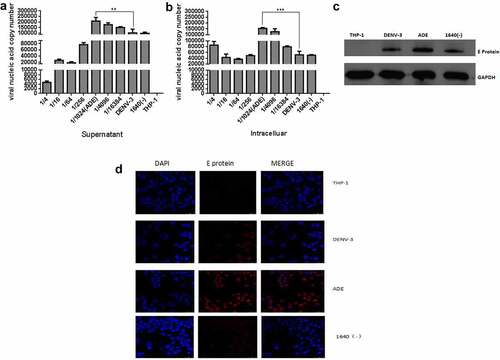
Table 1. Transcriptome sequencing data quality assessment
Figure 2. Transcriptome sequencing data quality assessment
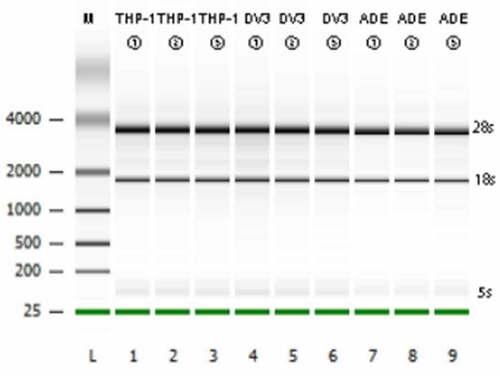
Figure 3. DE circRNAs expression profile
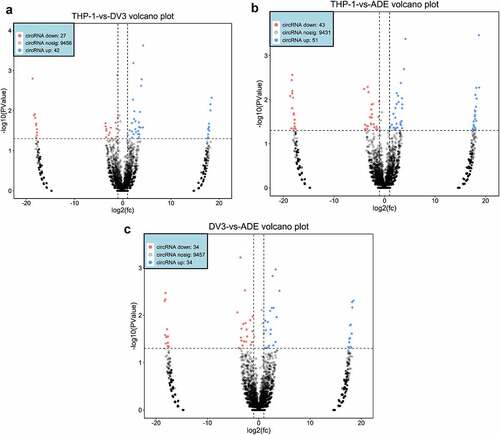
Figure 4. Differentially expressed mRNAs and miRNAs profiles
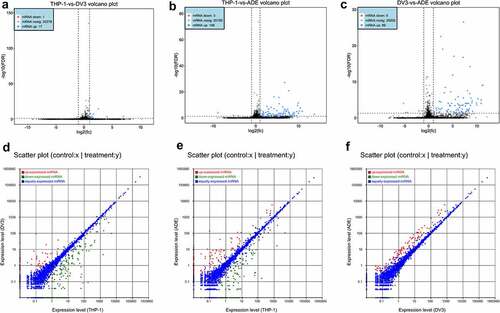
Figure 5. GO and KEGG analysis of DE RNAs
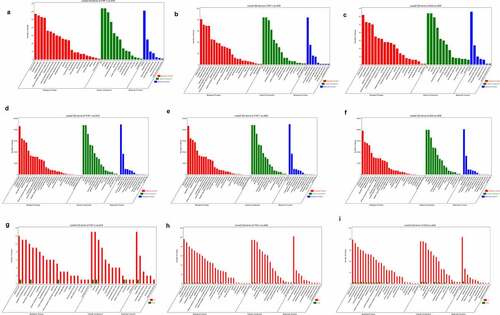
Figure 6. Quantitative real-time PCR analysis and Prediction of small peptide coding ability of circRNA encoded by host gene
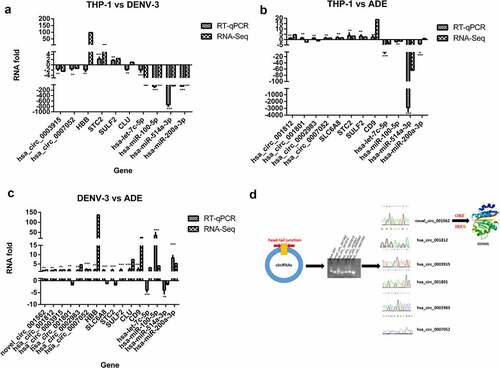
Figure 7. Differentially expressed CircRNAs- miRNAs- mRNAs interaction network
Supplemental Material
Download Zip (6.7 MB)Data availability statement
In this study, those data are available from the corresponding author based on reasonable requests.
