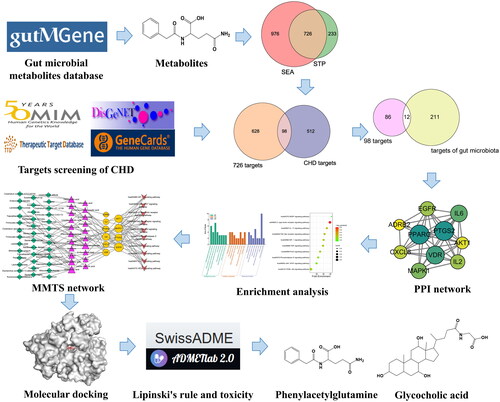
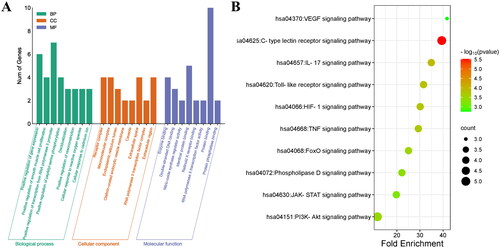
Figures & data
Figure 2. (A) The number of overlapping 726 targets between SEA and STP database. (B) The number of overlapping 98 targets between the 726 targets and CHD-related targets. (C) The number of the final overlapping 12 targets between the 98 targets and targets of gut microbiota. (D) The PPI network (10 nodes and 38 edges).
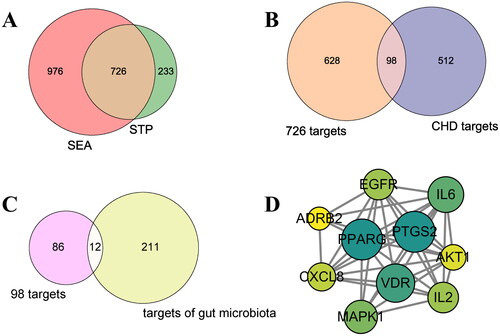
Figure 4. MMTS network (79 nodes and 118 edges). Green diamond: gut microbiota; pink triangle: gut microbial metabolites; yellow circle: targets; red quadrilateral: signalling pathways. The size of the node represents the number of relationships to the target.
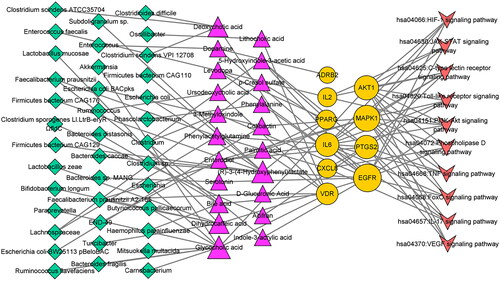
Table 1. Metabolites and core targets of major signalling pathways.
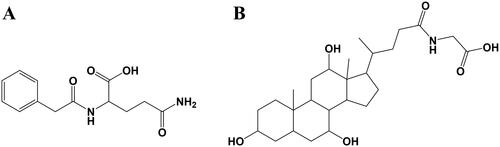
Figure 5. The molecular docking test on core targets-metabolites pairs. (A) Glycocholic acid-AKT1. (B) Colibactin-MAPK1. (C) Indole-3-acrylic acid-EGFR. (D) Phenylacetylglutamine-PTGS2.
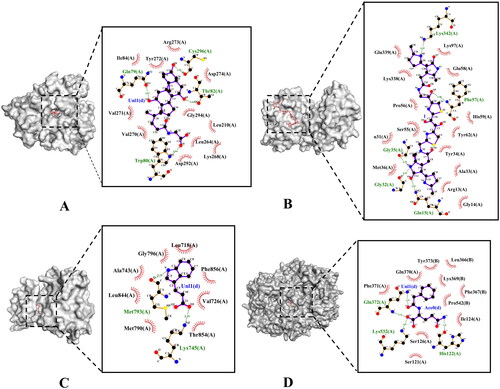
Table 2. Molecular docking results of metabolites and core targets.
Table 3. The physicochemical properties of the metabolites from gut microbiota.
Table 4. Toxicological properties of the metabolites from the gut microbiota.
Supplemental Material
Download MS Excel (66.6 KB)Data availability statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information Files).