Abstract
Sesquiterpene lactones (STLs) are bitter tasting plant specialized metabolites derived from farnesyl pyrophosphate (FPP) that contain a characteristic lactone ring. STLs can be found in many plant families that are distantly related to each other and outside the plant kingdom. They are especially prevalent in the plant families Apiaceae and Asteraceae, the latter being one of the largest plant families besides the Orchidaceae. The STL diversity is especially large in the Asteraceae, which made them an ideal object for chemosystematic studies in these species. Many STLs show a high bioactivity, for example as protective compounds against herbivory. STLs are also relevant for pharmaceutical applications, such as the treatment of malaria with artemisinin. Recent findings have dramatically changed our knowledge about the biosynthesis of STLs, as well as their developmental, spatial, and environmental regulation. This review intents to update the currently achieved progress in these aspects. With the advancement of genome editing tools such as CRISPR/Cas and the rapid acceleration of the speed of genome sequencing, even deeper insights into the biosynthesis, regulation, and enzyme evolution of STL can be expected in the future. Apart from their role as protective compounds, there may be a more subtle role of STL in regulatory processes of plants that will be discussed as well.
1. Introduction
The sesquiterpene lactones (STL) are a group of specialized plant metabolites with a 15-carbon backbone and a characteristic lactone ring. From the more than 11,000 reported sesquiterpenes more than 5,000 are STLs (Schmidt, Citation2006). Most of the reported STLs can be found in the Asteraceae (Rodriguez et al., Citation1976; Picman, Citation1986). STLs also occur in a few other angiosperm families such as Apiaceae, Magnoliaceae, and Illiciaceae, nonangiosperm land plants such as gymnosperms (ginkgo) and liverworts, but also in basidiomycetes and corals (Knoche et al., Citation1969; Magnusson et al., Citation1973; Bifulco et al., Citation1993; Huang et al., Citation2003). The plant lineages that produce STL are only distantly related, indicating several possible evolutionary origins of STL biosynthesis. The vast diversity of STLs, especially in the Asteraceae family, makes them an ideal target for chemosystematic, evolutionary and ecological investigations. STL can contain 5-membered γ-lactone rings, or 6-membered δ-lactone rings. The knowledge of the antimalarial effect of artemisinin (a δ-lactone) (23) that was dormant in ancient Chinese medical textbooks and re-discovered in the 1970s (Tu, Citation2016) is the most notable example for the pharmacological potential of STLs. In most other biologically active STLs the bioactivity is based on a highly reactive α-methylene-γ-lactone group (Kupchan et al., Citation1970; Schmidt, Citation1999). Reported biological activities of STLs include potential pharmaceutical applications as well as their roles in the natural environment of the producing plant. STLs are also appreciated as the bitter tasting ingredients of chicory, lettuce, and other Asteraceae species. In recent years, the amount of data on STLs has dramatically increased in the fields of (1) reported STL structures and spectroscopic data (https://mona.fiehnlab.ucdavis.edu), (2) elucidated STL biosynthesis genes (https://erda.dk/public/vgrid/PlantP450/), (3) transcriptome and whole genome sequences of STL-producing plants (https://www.ncbi.nlm.nih.gov/genome?term=Asteraceae) and (4) biological activities of STLs. For STL with a γ-lactone ring, biosynthetic pathways leading to all four possible ways for the formation of a lactone ring are known today. In addition, the decoration as well as the backbone conversion of costunolide (3), one of the key metabolites in STL biosynthesis, are understood. There are excellent reviews on the occurrence and chemosystematics of STL (Seaman Citation1982), their pharmacological applications (Ghantous et al., Citation2010), and their roles in protecting plants against herbivores, microbes, viruses and competitors (Picman, Citation1986; Padilla-Gonzalez et al., Citation2016). This review will shed light on some crucial aspects of STLs that have not yet been reviewed in detail: (1) the biosynthesis as well as the developmental, spatial, and environmental regulation of STL production and accumulation, (2) the opportunities for genomics and genome editing in STL research, (3) the role of STLs as potential signaling molecules in plants.
2. Structural diversity and Stereochemistry of sesquiterpene lactones
2.1. STL lactone rings and backbones
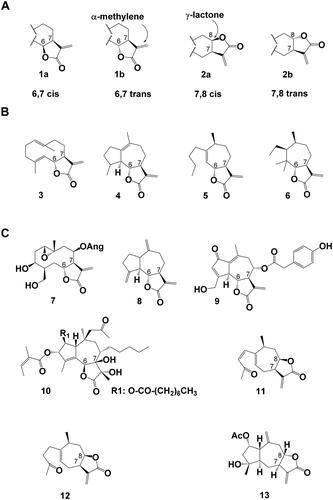
The chemical diversity of STL is based on a variety of backbone structures, different formation of the lactone ring and a plethora of decorations of the STL core backbones. Most STL are γ-lactones. That means that they are the product of an intramolecular esterification (lactonization) of a carboxy-group with a hydroxyl-group in the γ-position. The γ-lactones can be realized in four different ways—6,7 cis (1a) or trans (1b) and 7,8 cis (2a) or trans (2b) and contain in most cases an α-methylene moiety (). According to Seaman (Citation1982), the more than 30 STL backbones observed in the Asteraceae can be grouped into four complexity levels (derivable from the same number of modifications of the carbon skeleton) Examples for these backbones are germacranolides such as costunolide (3) for level I, guaianolide for level II (4), xanthanolide (5) for level III and 3,4-secoambrosanolide (6) backbones for level IV ().
Figure 1. Sesquiterpene Lactones (A) Stereochemistry of α-methylene-γ-lactone sesquiterpene lactones, (1a) 6,7 cis lactone, (1b) 6,7 trans lactone, (2a) 7,8 cis lactone (2b) 7,8 trans lactone. (B) Examples for sesquiterpene lactone backbones of increasing complexity levels as defined by Seaman, (Citation1982). (3) germacranolide (here: costunolide), (4) guaianolide, (5) xanthanolide, (6) 3,4-secoambrosanolide. (C) Examples for chemical STL diversity found in nature: (7) argophyllin B, (8) dehydrocostus lactone, (9) lactucopicrin, (10) thapsigargin, (11) 8-epi-xanthatin, (12) tomentosin, (13) 2α-acetoxy-inuviscolide. Ac: acetate, Ang: angelate.
2.2. Examples for STL diversity
Argophyllin B (7) () is a 6,7-trans germacranolide STL from sunflower trichomes. Dehydrocostus lactone (8) and lactucopicrin (9) are 6,7-trans and thapsigargin (10) is a 6,7-cis guaianolide STL. 8-epi-xanthatin (11) and tomentosin (12) are 7,8-cis xanthanolide STL that only differ in one double bond. 2α-acetoxy-inuviscolide (13) is a 7,8-trans guaianolide from Inula hupehensis. Costunolide (3), dehydrocostus lactone, tomentosin and 8-epi-xanthatin (3, 8, 11, and 12) are germination inducers of the parasitic plant Orobanche cumana and appear, like strigolactones, to have physiological roles in the plant. Thapsigargin (10) is the major toxic compound in Thapsia garganica roots and is a potential pro-drug for the treatment of prostate cancer. Argophyllin B and lactucopicrin (9) are defence compounds that can be found in the trichomes of sunflowers, or the laticifers of chicory respectively. The α-methylene present in most STL (3–9, 11–13) is often responsible for their bioactivity enabling Michael-type addition reactions (Michael, Citation1887) to biological nucleophiles such as the thiol group of cysteine (Cavallito and Haskell, Citation1945). However, in some STLs which lack the α-methylene group (e.g., dihydrohelenalin, artemisinin (23) and thapsigargin (10)), other functional groups such as a cyclopentenone ring or a peroxide group may account for the reported biological and pharmacological activities. Hydroxy groups decorating the backbone of the STL can be esterified to a variety of aliphatic and aromatic organic acids, as can be observed for compounds (7, 9, 10 and 13). These few examples show some of the complexity and diversity of STL found in nature raising the following questions: How are the different STL backbones produced? How are the γ-lactone rings with different configurations synthesized? Are there conserved biochemical pathways across several evolutionary lineages? What enables the stereospecific decorations of the backbones? Recent results have shed light on the biosynthesis of STL and begun to provide answers to these questions.
3. STL biosynthesis
3.1. Enzyme characterization
3.1.1. How to identify and characterize new enzymes in STL biosynthesis
Candidate enzymes for STL biosynthesis have been identified using comparative transcriptomics of different stages of secretory tissues (Göpfert et al., Citation2009; Liu et al., Citation2011, Citation2014) or genome sequences (De Bruyn et al., Citation2023) as well as phylogenetic analyses of P450 enzymes (). The genes for the biosynthesis of plant specialized metabolites are often found in genomic clusters (Nützmann and Osbourn, Citation2014). No such gene cluster for the biosynthesis of STLs was identified for a complete pathway, but there are hints to a gene cluster for higher oxidized STLs in sunflower and chicory (Frey et al., Citation2019; De Bruyn et al., Citation2023). The methods of choice for the identification and characterization of enzymes involved in the biosynthesis of STL have been summarized in detail before (Frey, Citation2020). Briefly, the options for STL enzyme characterization are: (1) Agrobacterium-mediated transient expression and the in vivo reconstruction of the metabolic pathway in Nicotiana benthamiana (Liu et al., Citation2011). (2) The in vivo reconstruction of the metabolic pathway in yeast (Saccharomyces cerevisiae)(Ro et al., Citation2006; Baek et al., Citation2021). (3) In vitro enzyme assays: Sesquiterpene synthases can be purified for in vitro assays from E. coli cultures (Göpfert et al., Citation2010), whereas the in vitro characterization of CYPs usually requires the preparation of yeast microsomes (Ikezawa et al., Citation2011). (4) Identification by loss-of-function mutations in planta (CRISPR/Cas for KLS and LCS in chicory). Other hosts for the heterologous production of STL include Nicotiana tabacum (Fuentes et al., Citation2016), the moss Physcomitrium patens (King et al., Citation2016) and E. coli (Chang et al., Citation2007). Depending on the expression and extraction system, rearrangements, and conjugation reactions can occur that need to be considered when analyzing the STL enzyme products. They include conjugation to glucose, cysteine, and glutathione as well as heat- and acid induced rearrangements (Frey, Citation2020).
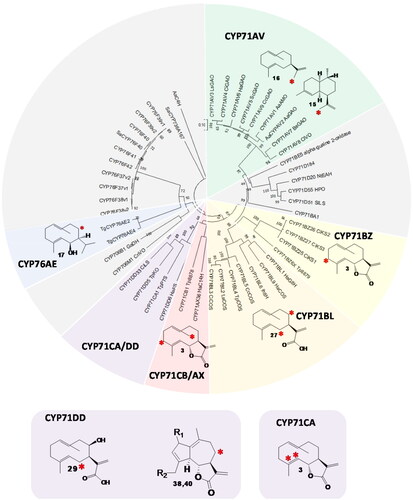
Figure 2. Phylogeny of cytochrome P450 enzymes (CYPs) that oxidize sesquiterpenes. Neighbour-joining tree with 500 bootstraps, outgroup: AaC4H, subfamilies with CYPs oxidizing sesquiterpenes but not involved in STL biosynthesis are colored grey. CYP subfamilies involved in STL biosynthesis are shown in green (CYP71AV), yellow (CYP71BZ), orange (CYP71BL), red (CYP71CB and AX), purple (CYP71CA and DD) and blue (CYP76AE). The positions of the oxidized carbons of the substrate are indicated by a red asterisk. (38): R1=R2=H, (40): R1=O, R2=OH.
3.1.2. Sesquiterpene synthases
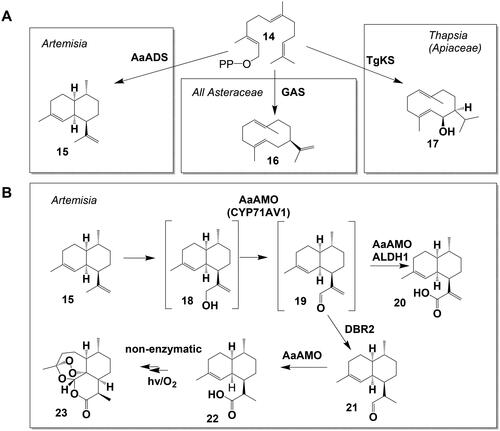
All STLs are derived from the universal precursor farnesyl pyrophosphate (FPP) (14) (). In the first step of STL biosynthesis, FPP (14) is converted by a sesquiterpene synthase (STS) to one or several sesquiterpene backbones. Most STL that have been elucidated to this date derive from the germacrene backbone of germacrene A (16), produced by a germacrene A synthase (GAS) (Bouwmeester et al., Citation2002; Nguyen et al., Citation2016). Two other STS, amorphadiene synthase (Mercke et al., Citation2000; Wallaart et al., Citation2001a) producing amorphadiene (15) and kunzeaol synthase (TgKS), producing kunzeaol (17) (Pickel et al., Citation2012) are involved in the generation of STL. Plant STS are often multi-product enzymes producing various sesquiterpene backbones. Many of these backbones can be found in the STL (Degenhardt et al., Citation2009). However, early biochemical models for the STL biosynthesis in the Asteraceae () suggested costunolide (3) as a common precursor for STL of many different backbones, requiring a conversion of the backbone after the formation of the lactone ring (Fischer et al., Citation1979; Seaman, Citation1982). Indeed, such backbone rearrangement after the formation of the lactone ring has been observed for the conversion of costunolide (3) (germacrene backbone) into kauniolide (37) (guaiane backbone) (Liu et al., Citation2018). Also, the pH dependent conversion from germacranolide to eudesmanolide backbone was observed in planta (Frey et al., Citation2018).
Figure 3. Sesquiterpene synthase reactions and artemisinin biosynthesis. (A) Sesquiterpene synthase reactions involved in STL biosynthesis pathways leading from farnesyl pyrophosphate (FPP, (14)), to amorphadiene (15), germacrene A (16) and kunzeaol (17). (B) Artemisinin biosynthesis from amorphadiene (15) via artemisinic aldehyde (19) to artemisinin (23) with the side product artemisinic acid (20).
3.1.3. Cytochrome P450 monooxygenase enzymes (CYPs)
Cytochrome P450 monooxygenases (P450s or CYPs) constitute a diverse and large enzyme superfamily, which typically cleave a molecule of molecular oxygen and insert one oxygen atom to the substrate, but they can also catalyze diverse reactions, including carbon cleavage and rearrangements. All 55 plant enzymes that oxidize sesquiterpenes belong to the CYP71 clan (Hansen et al., Citation2021) (https://erda.dk/public/vgrid/PlantP450/index.html, Supporting Data S1). Twenty-three published CYPs can oxidize sesquiterpenes but are not involved in sesquiterpene lactone biosynthesis. They are derived from the CYP families 71 (BE, D, Z), 76 (AE, AJ, F), 706 (M) and 736 (A) and oxidize various sesquiterpene backbones such as premnaspirodiene, drimenol or zingiberene (Takahashi et al., Citation2007; Henquet et al., Citation2017; Zabel et al., Citation2021). The 32 CYP enzymes reported to be involved in STL biosynthesis carry out the formation of hydroxy-, keto-, carboxy-, lactone- and epoxy-groups, as well as rearrangement reactions of the STL backbone (). Interestingly, CYPs involved in STL biosynthesis are very similar to each other: All CYPs involved in the biosynthesis of Asteraceae STL belong to the CYP71 family, one CYP involved in STL biosynthesis in the Apiaceae belongs to the CYP76 family (, ). Moreover, CYPs from the same subfamily often have similar substrates and carry out similar reactions in STL biosynthesis: Enzymes from the CYP71AV subfamily, for instance, carry out multi-step oxidations of olefinic sesquiterpene backbones (18, 25) and enzymes from the CYP71BL subfamily carry out a single hydroxylation of germacrene A acid (28) ().
Table 1. CYP enzymes involved in STL biosynthesis.
3.1.4. Other enzymes
Plant CYP enzymes require a redox partner in form of a cytochrome P450 reductase (CPR) and/or a cytochrome b5 (CYB5) that facilitates the supply of electrons required for the oxidation of their substrates. For example, the production of artemisinic acid in yeast was facilitated by co-expression of AaCPR and further improved through AaCYB5 (Ro et al., Citation2006; Paddon et al., Citation2013). A double bond reductase (DBR) (Zhang et al., Citation2008) was shown to be required for reduction of the exocyclic methylene group in artemisinin (23) formation. The esterification of hydroxylated STL backbones to organic acids requires enzymes such as BAHD acyltransferases, however, no such enzyme has been identified in STL biosynthesis pathways yet.
3.2. Biosynthetic pathways
3.2.1. Asteraceae
With over 25,000 species accounting for >10% of all Angiosperm plant species (Mandel et al., Citation2019), the Asteraceae is one of the largest plant families and contains a rich structural diversity of more than 5,000 STL. STL can be found in all larger Asteraceae subfamilies, except for the two basal lineages Barnadesioideae and Stifftioideae () and in most tribes of the Asteroideae, the by far largest subfamily of the Asteraceae ().
Table 2. STL in major subfamilies of the Asteraceae family and two outgroup families.
Table 3. STL in major tribes of the Asteroideae subfamily.
3.2.1.1. The biosynthesis of artemisinin
Due to its pharmacological potential, most notably as an antimalarial drug (Tu, Citation2016), the elucidation of artemisinin (23) formation was one of the first targets in the research on STL biosynthesis (). In the first step, farnesyl pyrophosphate (14) is converted to amorphadiene (15) by the Artemisia annua amorphadiene synthase (AaADS)(Wallaart et al., 2001b). The Artemisia annua amorphadiene oxidase (AaAMO, CYP71AV1) oxidizes amorphadiene (15) via artemisinic alcohol (18) and artemisinic aldehyde (19) to artemisinic acid (20) (Ro et al., Citation2006; Teoh et al., Citation2006). The Artemisinic aldehyde double-bond reductase (DBR2) (Zhang et al., Citation2008) converts the AMO intermediate artemisinic aldehyde (19) to dihydro-artemisinic aldehyde (21). Subsequently, the aldehyde dehydrogenase 1 (Aldh1) oxidizes dihydro-artemisinic aldehyde (21) to dihydro-artemisinic acid (22) (Teoh et al., Citation2009). Downstream dihydro-artemisinic acid, the last steps to the STL artemisinin (23) in Artemisia trichomes remain elusive but they are most likely the result of nonenzymatic photoreactions (Sy and Brown, Citation2002; Czechowski et al., Citation2016).
3.2.1.2. The biosynthesis of germacrene A acid
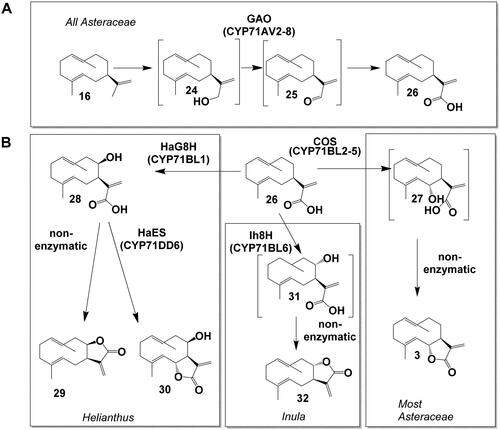
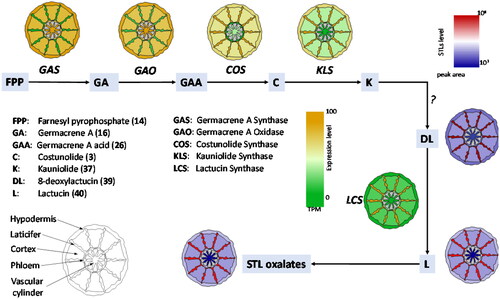
Germacrene A synthase (GAS) converts FPP (14) to the sesquiterpene backbone germacrene A (16). Germacrene A (16) is converted in a three-step oxidation, via germacrene A alcohol (24) and germacrene A aldehyde (25) to germacrene A acid (26) by the germacrene A oxidase (GAO, CYP71AV2-8) (Nguyen et al., Citation2010; Liu et al., Citation2011; Ramirez et al., Citation2013; Eljounaidi et al., Citation2014; De Bruyn et al., Citation2023) (). The three-step oxidation reaction of GAO resembles the reaction of AMO. Indeed, GAO homologs can be found in most lineages of the Asteraceae family and AMO is hypothesized to be derived from an ancestral GAO (Nguyen et al., Citation2010). Also, several GAOs from diverse Asteraceae plants accept amorphadiene as a substrate to produce artemisinic acid (Nguyen et al., Citation2010, Citation2019). Both GAS and GAO homologs were characterized throughout the Asteraceae family indicating that germacrene A acid is a key intermediate in the biosynthesis of Asteraceae STL. They were even found in the most ancestral Asteraceae lineage Barnadesioideae (Nguyen et al., Citation2010, Citation2016), from which no STL has been reported yet (). Germacrene A acid (26) has been observed to undergo acid-induced rearrangement reactions in unbuffered yeast media as well as in the Nicotiana benthamiana expression system, thereby forming the eudesmanolide backbone of costic acids (Nguyen et al., Citation2010; Frey et al., Citation2018). This rearrangement reaction might also occur in the natural cellular environment. Another enzyme from the CYP71AV family, the Cichorium intybus valencene oxidase (CiVO), carries out the same reactions as GAO and AMO, but can also convert (+)-valencene to +)-nootkatone (Cankar et al., (Citation2011).
3.2.1.3. The Lactonization of germacrene A acid
In the metabolic grid of STL biosynthesis, germacrene A acid (26) is a key branching point from which the biosynthesis of STL with different lactone configurations can occur (). CYP enzymes from the CYP71BL subfamily can oxidize GAA (26): Costunolide synthase (COS, CYP71BL2-5,7-11) converts GAA (26) to 6α-hydroxygermacrene A acid (27) that spontaneously lactonizes to the 6,7-trans lactone costunolide (3). This reaction occurs in many species and is conserved in most Asteraceae lineages (Ikezawa et al., Citation2011; Liu et al., Citation2011; Ramirez et al., Citation2013; Eljounaidi et al., Citation2014; Frey et al., Citation2020; De Bruyn et al., Citation2023). In Helianthus, GAA (26) can be oxidized by the Helianthus annuus germacrene A acid 8-hydroxylase (HaG8H, CYP71BL1) to 8β-hydroxy-germacrene A acid (28) (Ikezawa et al., Citation2011). Interestingly, when the expression system was changed from yeast to a plant, N. benthamiana, a subsequent lactonization to the 7,8-cis lactone inunolide (29) was observed (Frey et al., Citation2018). In Inula, GAA (26) can be oxidized by the nonstereoselective Inula hupehensis 8-hydroxylase (Ih8H, CYP71BL6) to either 8β-hydroxy-germacrene A acid (28) or 8α-hydroxygermacrene A acid (31), of which the latter spontaneously lactonizes to the 7,8-trans lactone epi-inunolide (32). Epi-inunolide could be the precursor of other 7,8-trans lactones such as 2α-acetoxy-inuviscolide (13) in Inula hupehensis (Gou et al., Citation2017). In sunflower capitate glandular trichomes, an alternative route to 6,7-trans lactones is realized by the Helianthus annuus eupatolide synthase (HaES, CYP71DD6), that converts 8β-hydroxy-germacrene A acid (28) to eupatolide (30) (Frey et al., Citation2018).
3.2.1.4. Oxidation reactions of costunolide
Costunolide (3) has been proposed to be a central precursor of the many STL with a 6,7-trans stereochemistry (). The H. annuus costunolide 14-hydroxylase (HaC14H, CYP71AX36) converts costunolide to 14-hydroxycostunolide (33), but also converts small amounts of parthenolide (34) to 14-hydroxyparthenolide (Frey et al., Citation2019). The Tanacetum parthenium parthenolide synthase (TpTPS, CYP71CA1) catalyzes the epoxidation of costunolide (3) to parthenolide (34) (Liu et al., Citation2014). A 3β-hydroxylase from Tanacetum (Tp8878, CYP71CB1) can introduce a 3β-hydroxy group to both costunolide and parthenolide (34), yielding either 3β-hydroxycostunolide (35) or 3β-hydroxyparthenolide (36) (Liu et al., Citation2014).
Figure 5. Biosynthesis pathways downstream costunolide in Asteraceae and STL biosynthesis in Apiaceae. (A) Conversion of costunolide (3) to 14-hydroxycostunolide (33) and 3β-hydroxycostunolide (35) (via hydroxylation), to parthenolide (34) (via epoxidation) and kauniolide (37) (via backbone rearrangement). (B) Oxidation of kauniolide (37) to lactucin (40). C Conversion of kunzeaol (17) to epi-dihydrocostunolide (41).
3.2.1.5. Biosynthesis of kauniolide and lactucin
The kauniolide synthase (KLS, CYP71BZ6X, 25-27) converts costunolide (3) to 3β-hydroxycostunolide (35), similar to CYP71CB1 (Tp8878) (). However, in the case of KLS, 3β-hydroxycostunolide (35) is only an intermediate, and the hydroxylation at C3 is followed by a rearrangement reaction to the guaianolide backbone of kauniolide (37) (Liu et al., Citation2018; Cankar et al., Citation2022). A similar mechanism could also lead to the formation of the kauniolide isomer dehydrocostus lactone (8). Kauniolide synthases were characterized from Tanacetum and Cichorium, and it is expected that KLS homologs are responsible for the conversion from germacrene to guaiane backbone in other Asteraceae lineages as well. The Tanacetum parthenium kauniolide oxidase (TpKO, CYP71DD5) can introduce a hydroxy group to kauniolide (37) (), but the position of the hydroxylation in this hydroxykauniolide enzyme product (38) is not known (Liu et al., Citation2018). Following the conversion of kauniolide (37) to 8-deoxylactucin (39) by yet unknown enzymatic steps, 8-deoxylactucin (39) is oxidized to lactucin (40) by the Cichorium intybus lactucin synthase (CiLCS, CYP71DD33) in chicory (Cankar et al., Citation2023). The generation of the 8α-hydroxy group enables the esterification to the side chain required for the biosynthesis of lactucopicrin (9). The biosynthesis of 8-epi-xanthatin (11) and tomentosin (12) is still enigmatic; one hypothesis could be that kauniolide synthase can also convert inunolide (30) to a guaianolide, and there is a subsequent conversion to the xanthanolide backbone by an opening of the C5 ring.
3.2.2. Other plant families
The occurrence of STLs in distant plant lineages raises the question of the evolutionary origin of STL biosynthesis in these clades, and it is possible, that the biosynthesis of STL has evolved independently in several plant lineages. Thus, structurally different CYPs may have been recruited for STL biosynthesis in these lineages. When looking for CYPs involved in STL biosynthesis outside the Asteraceae family, it makes sense to compare candidate sequences to all CYPs reported to oxidize sesquiterpenes (, clades with grey color, Supporting Data S1) (Andersen et al., Citation2017). The conversion of kunzeaol (17) to epi-dihydrocostunolide (41) in Thapsia garganica (Apiaceae) (is the only so far reported enzymatic reaction outside the Asteraceae (Andersen et al., Citation2017) () and the only example where a CYP enzyme from a family other than CYP71 is involved. Interestingly, the formation of a 6β-hydroxyl-group (by water quenching of the terpene synthase reaction) precedes the formation of the acid group that is then followed by the formation of a 6,7-cis lactone. Similar to AMO and GAO (CYP71AV1-8), the C12 carbon is oxidized in a three-step oxidation reaction. The product epi-dihydrocostunolide (41) is a likely precursor in the biosynthesis of thapsigargin (10) that requires a multitude of additional enzymatic reactions. Andersen et al. (Citation2019) found a similar CYP enzyme, TlCYP76AE4, that introduces an 8α-hydroxy group to epi-kunzeaol to yield tovarol. Outside of the Asteraceae and Apiaceae plant families, to the best of our knowledge, there is no report for a species where the biosynthesis of STL was elucidated.
4. Accumulation of STL and modulation of STL biosynthesis
Given the reactivity of STL, it is not surprising that their accumulation has often been associated with secretory structures, such as oil bodies - the most ancient secretory structures in land plants (Lange, Citation2015) - resin ducts, laticifers, and glandular trichomes. Occurrence of STL has for instance been observed in oil bodies of liverworts (He et al., Citation2013) and resin ducts of Apiaceae (Andersen et al., Citation2017). In this review, we will focus on STL in glandular trichomes and laticifers in the Asteraceae family, that have been thoroughly studied over the last decades and that can be found in many Asteraceae lineages (). These secretory structures, that often play a role in plant defence, can be veritable STL production factories and show extremely high local accumulation of STL. In the glandular trichomes, we now have a comprehensive understanding of how their development is connected to the development of the leaf or flower (detailed in chapter 4.1. and ). In the laticifers, the spatial distribution of STL production and gene expression across different tissue types has been investigated in detail in recent years (detailed in chapter 4.2. and ). We will also discuss the occurrence of STL outside of secretory tissues. The accumulation of STL in these tissues is lower than in secretory structures, by several orders of magnitude, indicating a more subtle role that is not fully understood yet (detailed in chapters 4.3. and 6).
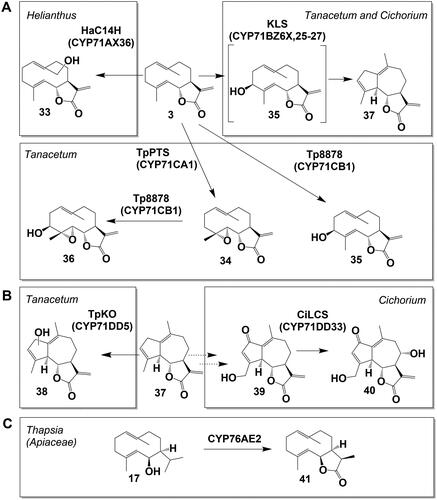
Figure 6. Helianthus annuus (sunflower) as a model for the developmental regulation of sesquiterpene lactone (STL) biosynthesis in (biseriate) capitate glandular trichomes (CGT) of Asteraceae. Scheme of CGT in (A) pre-secretory and (B) in secretory stage, respectively. Fully developed trichomes consist of 5-6 biseriate layers of stalk cells and 2 layers of secretory cells. STL are sequestered between cell wall and cuticle of secretory cells forming an extracellular glandular globe. (C) Scheme of (I) disk floret development in the capitulum. (II) Development of CGT on anthers, (III) biosynthetic STL gene expression (yellow/green gradient) in secretory cells and (IV) STL accumulation (red/blue gradient) in the glandular globe are tightly co-regulated. When the outermost disk floret opens, CGTs of the outer half of disk floret rows are already in the post-secretory stage while the innermost disk florets are still in the pre-secretory stage. (Göpfert et al. Citation2005, Citation2009). Note: The scheme of a glandular trichome in is modified from Spring et al. (Citation2020), The scheme of the cross section of a capitulum (flower head) in is modified from Göpfert et al. (Citation2005). Both original figures were published under the Creative Commons Attribution License.
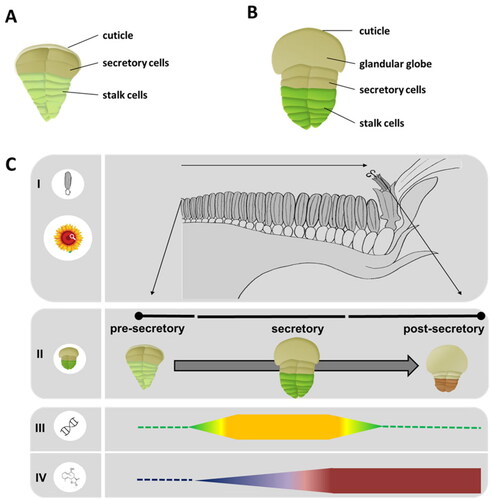
Figure 7. Schematic representation of the spatial distribution of gene expression (yellow/green gradient map) and STL accumulation (red/blue gradient map) over a schematic root structure from chicory based on data of previous publications (Bogdanović et al., Citation2019; Cankar et al., Citation2022; Citation2023; Vahabi et al., Citation2024). Genes involved in early steps of STL biosynthesis are expressed outside of the laticifers. Downstream of costunolide the expression pattern of STL biosynthesis genes is higher in the latex relative to other tissues (yellow/green gradient map). The accumulation of STLs is higher in the laticifers (red/blue gradient map) than in the other root tissues.
4.1. Glandular trichomes
4.1.1. Occurrence of STL in capitate glandular trichomes (CGT)
After more than half a century of research, most lineages of the Asteraceae have been investigated both for the presence of STLs as well as for the presence of glandular trichomes (, ). A large systematic screening for STL in Asteraceae started in the 1960s with the emergence of improved chromatographic and spectroscopic techniques. Within only two decades, hundreds of species were investigated, and more than 1,300 different STL compounds could be identified and were used for chemosystematic interpretation (Seaman, Citation1982). Amazing amounts of up to 5% of the dry weight of extracted plant material (Picman, Citation1986) were gained from solvent washings of aerial plant parts, hence suggesting their origin from secretory tissues located on the surface such as trichomes observed in Parthenium hysterophorus (Rodriguez et al., Citation1976) and many other taxa. Glandular trichomes are very common in Asteraceae and various different morphological types exist in this huge plant family (Werker, Citation2000). Not all of them are involved in the production, secretion, or storage of STL. Among the most frequent types found on Asteracean plant surfaces are uniseriate multicellular glandular trichomes which, for instance, produce mono-and sesquiterpenes (Aschenbrenner et al., Citation2013), and biseriate capitate glandular trichomes (CGTs), which were shown to synthesize and store STL (Spring et al., Citation1987; Duke et al., Citation1994), sometimes in combination with flavonoids (Schilling, Citation1983; Göpfert et al., Citation2006), myo-inositol (Spring et al., Citation1994) or other compounds. CGTs mostly occur on aerial parts of Asteraceae, often on leaves and flowers, less frequently on stems, and very seldom on roots as reported from Chrysolaena (Appezzato-da-Glória et al., Citation2012). However, the occurrence of CGTs appears to be sporadic, even among closely related taxa. This sometimes impeded the detection of STL in former studies but can be overcome by close microscopic observation prior to selective extractions of CGT-bearing plant parts (Spring, Citation2000). Thus, several previous negative reports on the presence of STL in Helianthus species could be revised after detection and extraction of CGTs solely located on anther appendages (Spring and Schilling, Citation1991). On the other hand, a recent review on Asteraceae mentioned the lack of glandular trichomes in 24 of the 35 investigated tribes (Martínez-Quezada et al., Citation2023), implying that other secretory structures such as canals, cavities or laticifers could be alternative sources if STL could be found in these taxa. Four of the most basal tribes (Barnadesieae, Hecastocleideae, Hyalideae and Stifftieae) seem to lack any type of secretory structures and so far have not been reported to contain any STL. Interestingly, homologous genes for germacrene A acid biosynthesis (GAS and GAO) have been identified in Barnadesioideae (Nguyen et al., Citation2010, Citation2016, Citation2019), indicating that the pathway down to germacrene A acid is evolutionary basal in Asteraceae and independent from the formation of secretory structures (). In the Asteroideae, the evolutionarily most advanced and by far largest subfamily of the Asteraceae, most larger tribes contain CGTs and STLs (). One exception are the Tageteae, which contain neither STL nor biseriate CGT (Martínez-Quezada et al., Citation2023).
4.1.2. Developmental regulation of STL production and gene expression in trichomes
The formation of capitate glandular trichomes (CGT) usually starts at a very early developmental stage of the plant organ, be it a leaf or the flower. In the presecretory glandular trichome the cuticle is still attached to the cell wall of the secretory cell (). The CGT formation is often completed before the leaf or flower has fully emerged (Werker, Citation2000). STL-producing biseriate CGT are composed of stalk cells, secretory cells, and a glandular globe, that accumulates the STLs (). Fully developed trichomes consist of 5-6 biseriate layers of stalk cells and 2 layers of secretory cells. STLs are sequestered between the cell wall and the cuticle of secretory cells forming an extracellular glandular globe (). The developmental process of biseriate CGT in Asteraceae has been documented in detail particularly in two plants, Artemisia annua (Duke and Paul, Citation1993) and sunflower (Helianthus annuus) (Spring and Bienert, Citation1987; Aschenbrenner et al., Citation2015). In sunflower, the development of the trichomes as well as the induction of STL genes and the accumulation of STLs therein is tightly correlated with the development of the flower (Göpfert et al., Citation2005, Citation2009) (). This correlation was also observed for the development of trichomes on leaf primordia (Aschenbrenner et al., Citation2015; Frey et al., Citation2018). A similar time course for the expression of enzymes involved in STL biosynthesis was reported for flower heads of Tanacetum cinerariifolium by Ramirez et al. (Citation2013) and in Tanacetum parthenium by Liu et al. (Citation2014). Using a laser microdissection pressure catapulting technique, Olsson et al. (Citation2009), located the site of artemisinin (23) biosynthesis in A. annua by tracing the activity of three cytochrome P450 enzymes involved in the conversion of FPP to artemisinin (23) in the two apical cells of biseriate CGT. This was later amended by Olofsson et al. (Citation2012) who detected gene expression of STL biosynthesis genes in sub-apical cells as well. The latter is in accordance with fluorescence immunolabelling experiments which showed the presence of germacrene A monooxygenase (HaGAO), a key enzyme of sunflower STL biosynthesis in secretory as well as in stalk cells (Amrehn et al., Citation2016). Moreover, these experiments showed that enzyme occurrence temporally correlates with the separation of the cuticle from the cell wall of the secretory cells and the beginning of metabolite sequestration. Furthermore, immunogold labeling in secretory and stalk cells of sunflower CGT showed that HaGAO is localized on the smooth endoplasmic reticulum (Amrehn et al., Citation2016). Extensive accumulation of smooth ER in the peripheral plasma of secretory and upper stalk cells suggests participation in vesicular transport of STL into the subcuticular space of trichomes. As proposed by Fahn (Citation2000), this appears to be a common mode for the secretion of lipophilic compounds in glandular trichomes of many plant families and ensures the elimination of highly reactive compounds from the cytoplasm.
4.2. Laticifers
4.2.1. Occurrence of STL in laticifers
Laticifers are long, tube-like cells that run through plant tissues and produce and store latex, a milky cytoplasmic liquid that contains various specialized metabolites. Laticifers can be classified into the following two types: articulated, which are composed of a series of cells joined together, and nonarticulated, which consist of one long coenocytic cell (Hagel et al., Citation2008). This network of elongated cells can be found in the proximity of vascular bundles of all plant tissues. In chicory, articulated laticifers compose an anastomosing network that is tightly associated with the phloem in the vascular bundles (Vertrees and Mahlberg, Citation1978). Laticifers can be found in many plant genera of distantly related plant lineages, such as Euphorbia, Papaver and Ficus (Hagel et al., Citation2008). A wide range of plant specialized metabolites, such as benzylisoquinoline alkaloids in opium poppy (Desgagné-Penix and Facchini, Citation2012) or phorbol esters in Euphorbiaceae (Tostes et al., Citation2021), can be produced in laticifers; in the Asteraceae they accumulate mainly natural rubber (cis-1,4-polyisoprene) and STLs (Hagel et al., Citation2008; Kwon et al., Citation2022, Citation2023). The occurrence of laticifers has been reported for 1,800 Asteraceae species (Gutiérrez and Luna, Citation2013) that all belong to the four tribes of the Cichorioideae subfamily: Arctotideae, Cichorieae, Liabeae, and Vernonieae (Martínez-Quezada et al., Citation2023) (). In chicory roots, lactucin, 8-deoxylactucin, lactucopicrin (9, 39-40), and their oxalate conjugates are the major STLs that accumulate primarily in the latex () (Cankar et al., Citation2023). The STL oxalates are unstable and may on decomposition lead to the accumulation of oxalic acid in the latex released by leaf damage which may contribute to the sensory and antifeedant properties (Sessa et al., Citation2000). In the latex of Lactuca species STL are mainly found as oxalate and sulfate conjugates (Sessa et al., Citation2000).
4.2.2. Spatial and developmental regulation of STL production and gene expression in laticifers
In morphine biosynthesis in opium poppy, although laticifers are the site for morphine accumulation, other cells surrounding the laticifers (e.g., sieve elements and vascular parenchyma cells) contribute to the early parts of morphine biosynthesis (Bird et al., Citation2003; Weid et al., Citation2004; Onoyovwe et al., Citation2013). Similarly, the biosynthesis of STL is spatially separated in distinct cell types, and such a cellular compartmentalization has been best studied in chicory and lettuce. In chicory, the genes for the early biosynthetic steps CiGAS and CiGAO, that convert FPP to GAA (26) are expressed outside of the laticifers () (Bogdanović et al., Citation2019; Cankar et al., Citation2022; Vahabi et al., Citation2024). The genes for the late STL pathway, costunolide synthase (CiCOS) as well as the kauniolide synthase (CiKLS) and the lactucin synthase (CiLS) are expressed in the laticifers, where the accumulation of the STL lactucin, deoxylactucin and STL oxalates is observed () (Cankar et al., Citation2022; Citation2023; Vahabi et al., Citation2024). In lettuce, a molecular approach using LsGAS1/2 promoters fused to β-glucuronidase (GUS) provided clear evidence that LsGAS1/2 are not expressed in laticifers but expressed in the vascular parenchyma cells surrounding the laticifers (Kwon et al., Citation2022). This is a stark contrast to the same GUS assays using the promoters of natural rubber biosynthetic genes (i.e., cis-prenyltransferase (CPT3) and CPT-binding protein (CBP2)), in which exclusive expression of CPT and CBP is observed in the lettuce laticifers (Qu et al., Citation2015; Barnes et al., Citation2021). It has been a mystery how lettuce laticifers biosynthesize and accumulate high concentrations of both natural rubber and STLs that share the same precursor, isopentenyl pyrophosphate (IPP). It appears that lettuce laticifers avoid the competition for IPP between the two biochemical pathways for natural rubber and STL biosynthesis by recruiting adjacent parenchyma cells as a separate cell-factory to produce STL backbones from a distinct IPP pool (Kwon et al., Citation2022). STLs with their α-methylene-γ-lactone moiety are highly reactive compounds. Thus, the specific localization of STL pathway enzymes in the laticifers that are required for the formation of the lactone ring constitutes a strategy to reduce toxicity for the rest of plant. Validating this interpretation, knocking-out the chicory kauniolide synthase gene leads to the accumulation of costunolide, but mostly in forms conjugated to cysteine and glutathione that neutralize the reactive α-methylene group (Cankar et al., Citation2022). Thus, the compartmentalization in laticifers offers two advantages: preventing auto-toxicity and providing protection for the vascular tissues that they surround. Recently, major latex proteins (MLPs) in opium poppy laticifers have shown alkaloid-binding activities for sequestration and some simple catalysis (Ozber et al., Citation2022). It would be interesting to examine whether MLPs in chicory and lettuce laticifers, which are also highly expressed there, are also able to sequester reactive STLs.
4.3. Roots and inner tissues of seedlings
For long time, secretory tissues such as CGT, laticifers or resin ducts were thought to be the exclusive sites of production and accumulation of STL in the Asteraceae and this was in accordance with the suggested protective function of the compounds. Although interference of STL with auxin-induced plant growth in sunflower hypocotyls had been observed already in the 1960s (Shibaoka, Citation1961; Morimoto et al., Citation1966) indicating a possible additional physiological role of the compounds, the identification of STL in nonsecretory tissues failed due to the low concentrations and poorly sensitive detections methods. It was the detection of 8-epi-xanthatin (11) in phototropically stimulated hypocotyls (Yokotani-Tomita et al., Citation1997; Citation1999) and of three additional STLs in root exudates of sunflower (Joel et al., Citation2011; Raupp and Spring, Citation2013) which indicated the existence of further sources for the biosynthesis of STL in inner tissues of the plant. HPLC and MS analysis showed that the four compounds, costunolide (3), dehydrocostus lactone (8), 8-epi-xanthatin (11) and tomentosin (12) exist in micromolar concentrations in oil from dry seeds (Spring, Citation2021) and also can be extracted in low amounts from cotyledons, hypocotyls and roots of seedlings during the first days after germination (Spring et al., Citation2020). Interestingly, the identified compounds belong to three different skeletal types (germacranolides, guaianolides, xanthanolides), which are rare or not represented in the STL profile known from sunflower trichomes (mostly harboring heliangolides). Gene expression studies for the key enzymes HaG8H (essential for the synthesis of the xanthanolides tomentosin (12) and 8-epi-xanthatin (11)), and HaCOS (necessary for the germacranolide costunolide (3) and the guaianolide dehydrocostus lactone (8)) confirmed the early existence of two independent pathways of the endogenous STL in these organs (Spring et al., Citation2020).
4.4. Effect of external factors on STL production
Research indicates that plant STL accumulation is primarily driven by developmental factors. However, both abiotic and biotic factors can impact gene expression associated with STL biosynthesis and the accumulation of specific metabolites with protective or signaling roles. Therefore, understanding the complex interplay between these abiotic and biotic factors is crucial for uncovering the ecological and physiological importance of STL accumulation in plants, which holds implications for both agricultural and pharmaceutical industries.
4.4.1. Abiotic factors
Among abiotic factors, the accumulation of STLs in plants is mainly influenced by light intensity, temperature, and soil composition (Spring et al., Citation1986; Scavo et al., Citation2020; Greinwald et al., Citation2022). An early investigation from the mid-1980s provided the first valuable insights into this phenomenon (Spring et al., Citation1986). High-intensity light significantly increased niveusin C and 15-hydroxy-3-dehydrodesoxyfruticin accumulation in sunflower seedlings compared to low-intensity light (Spring et al., Citation1986). However, in field conditions STL accumulation can also be induced by shading: A recent study with Cynara cardunculus L. plants subjected to 60% shading in the field showed increased STL accumulation compared to control plants in natural light (Scavo et al., Citation2020). Additionally, maintaining illumination during chicory (Cichorium intybus L.) storage led to a significant rise in STL levels and bitterness (Wulfkuehler et al., Citation2014). These studies collectively highlight that lower-than-natural light exposure leads to higher STL accumulation, contrasting with the impact of light intensity. Besides light treatments, soil composition, temperature, and altitude strongly correlate with STL accumulation (Perry et al., Citation2009; Seemann et al., Citation2010; Lu et al., Citation2013; Greinwald et al., Citation2022). Transcriptome studies on Artemisia annua revealed that key genes for artemisinin (23) biosynthesis increased over two-fold with high-temperature exposure (40 °C), leading to increased artemisinin contents (Lu et al., Citation2013). Studies on flower heads of Arnica montana L. concluded that the response of STL to climatic factors is compound specific, and that some of them, such as 6-O-isovalerylhelenalin, exhibit more pronounced and statistically significant correlations with specific climatic variables (Seemann et al., Citation2010). A follow-up study on different soil types in the same region and altitude for Arnica montana L. showed that habitat conditions and abiotic and biotic soil parameters influence STL accumulation in flower heads (Greinwald et al., Citation2022). These findings highlight the variable responses of different STLs to climatic conditions and the significant influence of abiotic factors on STLs biosynthesis.
Also, the application of exogenous substances can modulate the expression of STL genes in different plant species (Pu et al., Citation2009; Maes et al., Citation2011; Majdi et al., Citation2013, Citation2015). Plant hormones like jasmonic acid, abscisic acid, and salicylic acid increase the artemisinin (23) content in A. annua (Pu et al., Citation2009; Maes et al., Citation2011), parthenolide (34) in feverfew (Tanacetum parthenium) (Majdi et al., Citation2015), and guaianolide STLs in Cichorium intybus (via methyl jasmonate) (De Bruyn et al., Citation2023). Transcriptome studies revealed that these phytohormones share a similar mechanism of STL induction, by upregulating key genes in STLs biosynthesis. Similar results have been obtained by treatment with other substances. Feverfew (T. parthenium) plants treated with 2,4-dichlorophenoxyacetic acid showed a significant increase in parthenolide (34) and costunolide (3) (Majdi et al., Citation2013). Microbial fertilizers (EM Aktiv and Vital Tricho) boost lactucopicrin (9) and other STLs in lettuce (Lactuca sativa) (Stojanović et al., Citation2023). While most studies focus on germination and growth bioassays in controlled laboratory conditions, field investigations aiming to determine the impact of environmental conditions on STL biosynthesis and accumulation in nature are limited (Scavo et al., Citation2020). Prior research by Padilla-González et al. (Citation2019, Citation2020) explored field conditions over six months for yacon (Smallanthus sonchifolius) but could not draw definitive conclusions about STLs due to the consistent levels seen from the first collection. Hence, additional field studies or simulations mimicking natural conditions are essential to understand how the environment regulates STL biosynthesis. These studies should encompass natural light, temperature variations, and soil compositions during early seedling and leaf development, to answer long-standing questions like: How does the environment regulate the STL biosynthesis and accumulation in nature? Is it possible to select optimal locations and seasons for maximizing the production of biologically important STL?
4.4.2. Biotic factors
Another key determinant for the accumulation of STL in plants lies in the intricate interplay of biotic factors. Interactions with herbivores and symbiotic microorganisms correlate with increased accumulation of STL in plants (Rozpądek et al., Citation2014; Huber et al., Citation2016). In lettuce, incompatible interaction (successful defence) with the pathogen Bremia lactucae rapidly induces the biosynthesis of lettucenin A (a guaianolide STL) in infected tissues, while no such induction of lettucenin A could be detected in compatible (disease-causing) interaction (Bennett et al., Citation1994). Lettucenin A exhibited potent antimicrobial activities against B. lactucae and P. syringae at low µM concentrations, implying its defensive role. Increased Melolontha melolontha infestation in Taraxacum officinale correlated with higher taraxinic acid β-D-glucopyranosyl levels in root latex (Huber et al., Citation2016). The discovery of heritable variations in the levels of taraxinic acid β-D-glucopyranosyl within T. officinale populations by Huber and coworkers (2016) implies that STL concentration is subject to M. melolontha-imposed divergent selection pressures. The stereochemistry of the STL ring also influences herbivore resistance, with cis-fused STL-producing plants experiencing more damage than trans-fused lactone producers (Ahern and Whitney, Citation2014). Apart from their role in herbivore response, STLs serve as signaling compounds in the rhizosphere, facilitating communication among plants, soil microorganisms, and parasitic weeds (Macías et al., Citation1996; Macías et al., Citation1999; Andolfi et al., Citation2013; Padilla-Gonzalez et al., Citation2016). For example, chicory root STL accumulation correlates positively with arbuscular mycorrhiza fungal associations (Rozpądek et al., Citation2014).
4.5. Manipulation of STL pathway in planta
The manipulation of the biosynthetic pathway of STL in Asteraceae plants has been documented in species that produce STLs in both latex and trichomes. In Taraxacum officinale the silencing of the germacrene A synthase gene ToGAS1 by RNAi resulted in 90% reduction of taraxinic acid β-D-glucopyranosyl ester levels and a significant increase in M. melolontha feeding, clearly demonstrating the role of a latex STL in plant fitness under herbivore attack (Huber et al., Citation2016). In chicory, prevention of STL accumulation in latex is relevant for inulin extraction. Inulin is a prebiotic and sweetener commercially extracted from taproots of industrial chicory (Roberfroid, Citation2007). During the extraction procedure the bitter tasting STLs need to be removed, resulting in additional processing costs (Hingsamer et al., Citation2022). Additionally, manipulation of bitter STL content is relevant in the case that the reduction of bitterness in leaf chicory varieties is desired (e.g., Belgian endive, radicchio). The first step of the STL pathway in chicory is catalyzed by the CiGAS enzyme which is encoded by 4 genes in the chicory genome (Bogdanović et al., Citation2019). Silencing of the CiGAS gene by artificial microRNAs in chicory resulted in reduction of CiGAS expression and decreased STL accumulation (Bogdanovic, 2020). Using CRISPR/Cas genome editing all 4 gene copies of CiGAS were interrupted successfully, resulting in elimination of STL production of chicory (Cankar et al., Citation2021). In these lines, squalene accumulation was observed, indicating that the FPP precursor pool was at least partially channeled to the production of this triterpene and phytosterol precursor which is normally not found to be accumulated in chicory taproots (Cankar et al., Citation2021). To reduce bitterness by inactivating downstream biosynthetic enzymes, a number of CYP71-clan enzymes were simultaneously targeted by CRISPR/Cas genome editing (De Bruyn et al., Citation2023). Elimination of STLs was achieved by simultaneous interruption of the three CiKLS paralogs, while other mutants showed shifts in STL compound concentrations, indicating that mutation effects of single copies of genes may be masked due to activity of intact paralogs. Additionally, gene editing of several genes encoding CYPs involved in the chicory STL pathway was carried out, with the intention of accumulating medicinal terpenes in chicory taproots. Thus, knocking-out the three copies of CiKLS genes resulted in accumulation of costunolide (3) in taproots that normally does not accumulate in this tissue (Cankar et al., Citation2022). Next to free costunolide (3), accumulation of costunolide conjugated to cysteine and glutathione was observed, presumably due to the reactivity of the α-methylene-γ-lactone moiety of costunolide. To specifically accumulate the anti-inflammatory 8-deoxylactucin (39) the activity of the lactucin synthase CiLCS was interrupted by genome editing (Cankar et al., Citation2023) of the CiLCS gene. This resulted in interruption of the lactucin (40) and lactucopicrin (9) production and increased the accumulation of 8-deoxylactucin (39) and its derivatives. Interestingly, in this case no conjugation was observed, presumably due to the localization of this endogenous STL in the latex. These examples show that silencing and genome editing approaches are useful to identify STL biosynthetic genes, eliminate STL biosynthesis, change latex STL chemotypes and enable study the role of latex STLs in plant defence. Regarding manipulation of the terpene pathway in trichomes, large efforts were dedicated to the engineering of increased production of artemisinin (23) in glandular trichomes of Artemisia annua. The approaches taken include targeting transgene expression to subcellular compartments, the overexpression of upper pathway genes (e.g. HMGR, FPS), overexpression of the genes for key biosynthetic enzymes (e.g. ADS, AMO, DBR2), downregulation of genes for competitive pathway enzymes (e.g. squalene synthase, copalyl diphosphate synthase), overexpression of transcription factor genes (e.g. AaWRKY, AaERF, AaORA, AaMyc2, AaNAC1) and other genes indirectly stimulating secondary metabolite production, such as for example the Agrobacterium rol genes, as previously reviewed (Majdi et al., Citation2016; Zakariya et al., Citation2023). By now, over 30 distinct transcription factors are reported to independently regulate artemisinin biosynthesis in Artemisia trichomes (see in Zakariya et al., (Citation2023), which might be unrealistic and should be taken with caution. In addition, the approaches can be combined to simultaneously overexpress transcription factors and biosynthetic enzymes (Hassani et al., Citation2023). Most successful engineering approaches yielding artemisinin (23) at more than 20 mg/g (DW) were achieved by overexpression of fused FPS and ADS (Han et al., Citation2016), downregulation of SQS (Zhang Citation2009) and overexpression of the transcription factor AcNAC1 (Lv et al., Citation2016). Next to overexpression and silencing of genes, genome editing may in the future provide a new tool for engineering of trichome STL content by interrupting genes or changing promoters of endogenous transcription factors and biosynthetic genes. However, some Asteraceae species such as sunflower are recalcitrant to in vitro culture and regeneration which may pose a limitation in the application of new plant breeding techniques and additional efforts are needed to develop efficient protocols.
5. Genomic organization of STL production
The pace of genome sequencing in the Asteraceae has accelerated in the past few years. As of January 2024, reference sequences for 50 Asteraceae taxa were publicly available (https://www.ncbi.nlm.nih.gov/genome?term=Asteraceae). These include a representative genome for most of the major subfamilies and tribes of the family (, Supporting ). Examples are sunflower (Helianthus annuus, Heliantheae tribe) and annual absinthe (Artemisia annua, Anthemideae tribe) for the Asteroideae subfamily, lettuce (Lactuca sativa) and chicory (Cichorium intybus) for the Cichorioideae subfamily, and globe artichoke (Cynara cardunculus) for the Carduoideae subfamily (Acquadro et al., Citation2017; Badouin et al., Citation2017; Fan et al., Citation2022; Liao et al., Citation2022; Shen et al., Citation2023a). In addition, a high-quality genome was recently published for the Goodeniaceae (Shen et al., Citation2023b), which is closely related to the Asteraceae and can be employed as an outgroup for comparative analyses. These many new genomes open up opportunities to rapidly identify new homologous enzymes encoded by STL pathway genes in other species, explore the genomic organization and regulation of STL biosynthesis, and perform targeted knock-out of genes in these species (Catania et al., Citation2018; Frey et al., Citation2019; Cankar et al., Citation2021, Citation2022). Genomic information may also permit inferences about the origin and evolution of STL genes, as well as possible impacts of innovations in STL biosynthesis on the diversification of the family. The genes and pathways underlying STL biosynthesis have been explored most thoroughly in Artemisia (Shen et al., Citation2018; Liao et al., Citation2022; Chen et al., Citation2023) and Cichorium (Fan et al., Citation2022; Zhang et al., Citation2022; Waegneer et al., Citation2023; Shen et al., Citation2023a). In Artemisia, the main interest has been in artemisinin (23) biosynthesis. In a draft assembly of the Artemisia annua genome, Shen et al. (Citation2018) identified 122 terpene synthase (TPS) genes, which is similar in number to those found in the H. annuus genome (Badouin et al., Citation2017), but much higher than that reported for most other plant species. For example, only 34 terpene synthases are found in the Arabidopsis thaliana genome. Phylogenetic analyses further showed that most Asteraceae TPS genes have arisen by gene and genome duplication after divergence from the lineages that contain the Solanaceae and Brassicaceae, represented by the model plants tomato and Arabidopsis, respectively. More recently, fully haplotype-resolved genomes were published for low and high artemisinin lines (Liao et al., Citation2022), which offered additional insights into the origins and diversity of genes contributing to artemisinin biosynthesis. These included the discovery of an additional 14 TPS genes, as well as differences in tandem duplication patterns between haplotypes and lines. Gene duplication was common throughout the artemisinin biosynthetic pathway, with at least one tandem or dispersed copy of all committed genes in all four haplotype genomes. However, only for ADS (amorpha-4,11-diene synthase), which encodes the enzyme that performs the first dedicated step in the artemisinin biosynthetic pathway, were there consistent differences in copy number between the high and low artemisinin lines. Resequencing of a diversity panel further demonstrated a strong correlation between ADS copy number and artemisinin (23). Interestingly, the loss of ADS homologs in a close relative (Artemisia argyi) appears to have resulted in the lack of artemisinin production in this species (Chen et al., Citation2023). These observations are consistent with an earlier RNAi experiment that reduced artemisinin (23) content by >95% by silencing all 12 ADS copies in the HAP variety of Artemisia annua (Catania et al.,Citation2018). Thus, ADS copy number could be targeted by breeding programs aimed at increasing artemisinin content. In Cichorium, high quality reference genomes have been developed for the two main crops in the genus: chicory (Cichorium intybus), which is cultivated for both its leaves and roots, and endive (Cichorium endivia), which is mainly grown for its leaves (Fan et al., Citation2022; Zhang et al., Citation2022; Waegneer et al., Citation2023; Shen et al., Citation2023a). Leaves of chicory and endive are popular in salads because of their bitter taste, which is caused by STLs. Thus, it is perhaps unsurprising that there has been a significant expansion of gene families associated with the terpenoid production in the Cichorioideae. This includes discovery of 30 HMGR genes, which exceeds that reported for most other representative plant species, both within and outside of the Asteraceae (Zhang et al., Citation2022; Shen et al., Citation2023a). Interestingly, twenty of the HMGR copies are found in a single cluster on chromosome 2; analyses of patterns of sequence divergence among paralogs indicate that they originated via a combination of whole genome duplications and tandem duplications. There also appears to be a recent burst of tandem duplications in a sub-clade of TPS genes (the TPS-a clade) that code for sesquiterpene synthases. Of 48 TPS genes in chicory, 26 (58%) belong to the TPS-a clade, of which 61% were generated by recent tandem duplications. Thus, Shen et al. (Citation2023a) suggested that duplication of HMGR and TPS-a genes may be responsible for sesquiterpene accumulation in chicory. Similarly, Zhang et al. (Citation2022) reported higher expression of several TPS-a paralogs (along with correlated sequence changes) in more bitter varieties of endives, possibly accounting for differences in STL accumulation between varieties. Of course, many gaps remain in our understanding of the origins, diversity, and evolution of genes contributing to the STL biosynthesis in the Asteraceae. Some of these could be addressed by analyzing existing genomes, which would allow us to fill in the gaps in , as well as to explore the genomic distribution of STL genes in the context of synteny relationships and whole genome duplications across the family. By comparison with the newly published genome for the Goodeniaceae (Shen et al., Citation2023b), it would be possible to determine whether homologs of GAS and GAO likely existed in the ancestor of the Asteraceae. The sequencing and assembly of new genomes would be useful as well, especially for Barnadesia, which represents the basal subfamily of the Asteraceae and Calyceraceae, which is sister to the Asteraceae and shares a paleotetraploid ancestor with it (Barker et al., Citation2016).
6. The biological role of sesquiterpene lactones in plant-plant interaction and in phototropism
STL play a crucial and well-described role in plant defense against herbivores, phytopathogens, and as allelochemicals in competition with other plants (Picman, Citation1986), which has attracted considerable attention for several decades. Interestingly, STLs are also involved in intra-plant physiological processes and in plant-plant interactions (Padilla-Gonzalez et al., Citation2016), that deserve a closer look: After the detection of the growth-inhibiting effect of STLs in the 1960s (Shibaoka, Citation1961; Morimoto et al., Citation1966), the way that STL affect growth was further investigated. The inhibition of auxin-induced elongation growth of Avena coleoptiles was observed, (Spring et al., Citation1981; Spring and Hager, Citation1982) as well as the STL induced increase of root growth in Phaseolus aureus seedlings (Kalsi et al., Citation1979; Singh et al., Citation1992). An involvement of STL in the auxin-mediated phototropism response was demonstrated by studies of Yokotani-Tomita et al. that showed a significantly higher accumulation of 8-epi-xanthatin (11) on the side of light exposure combined with inhibition of growth on the application side, leading to curvature and tropism toward that side (Yokotani-Tomita et al., Citation1997, Citation1999). In addition, external application of costunolide (3) resulted in directed growth of sunflower (H. annuus) hypocotyls toward the application side (Spring et al., Citation2020). Taken together, these results further support the hypothesis that STL may be involved in auxin-mediated growth regulation in plants. Although the inhibition of auxin-mediated growth has been known for a long time, the underlying mechanisms are so far not well understood. Based on experimental data and in silico analyses, some hypotheses can be formulated. STL growth-inhibition effect could be abolished by treatment with dithiothreitol, which suggests that the α-methylene-y-lactone moiety, as a characteristic structural element of STL, could be required for the functional activity (Spring and Hager, Citation1982). Exogenous application of the dehydrocostus lactone (8) to etiolated epicotyls of pea (Pisum sativum) showed an effect on the gene expression level of the auxin influx and efflux proteins PsAUX1 and PsPIN1, respectively (Toda et al., Citation2019). In addition, molecular docking analysis with the hydrolase Karrikin-insensitive 2 (KAI2) from sunflower revealed a high affinity for 8-epi-xanthatin (11) and tomentosin (12), providing a first indication that these STLs may be the elusive endogenous ligands of the karrikin receptors (Rahimi and Bouwmeester, Citation2021). KAI2 signaling regulates many developmental processes in plants, including germination, seedling photomorphogenesis, and root/root hair growth, and the endogenous ligand (KL) has not yet been identified (Varshney and Gutjahr, Citation2023). Recognition of endogenous STL by KAI2 could be a reasonable mechanism to integrate the growth inhibitory effects of STL. However, clear experimental evidence for STL as endogenous ligand or for other potential mechanisms for the integration of STL into plant growth regulation is still lacking and remains a worthwhile goal for further research. Besides their accumulation in trichomes and endogenous plant tissues, STLs are also known to be exuded from plant roots into the rhizosphere (Joel et al., Citation2011; Raupp and Spring, Citation2013; Wu et al., Citation2022). It is not yet clear, whether this exudation of STL is necessary for the plant itself, e.g. as a protective mechanism (antimicrobial/antifungal activity) or for a beneficial interaction (arbuscular mycorrhiza). However, the exudation of STL is used by parasitic plants for host recognition and specific germination only in the immediate vicinity of the host plants rhizosphere (Joel et al., Citation2011; Raupp and Spring, Citation2013; Wu et al., Citation2022). Four STLs have been identified in sunflower root exudates and have been shown to induce germination of the host-specific root-parasite Orobanche cumana (Joel et al., Citation2011; Raupp and Spring, Citation2013; Wu et al., Citation2022). Furthermore, the exuded STLs not only induced germination of O. cumana seeds, but also led to a chemotropic growth of the O. cumana germ tube toward the exogenous application side of the STL (Krupp et al., Citation2021). Similar experiments with the synthetic strigolactone GR24 only induced germination of O. cumana seeds, but failed to induce the chemotropic response of the corresponding germ tube (Krupp et al., Citation2021). Thus, these data indicate a STL-specific perception and chemotrophic growth response in O. cumana, but the underlying mechanisms are not yet known. It is tempting to speculate that STLs may be able to induce curvature and tropism toward the host plant by lateral inhibition of growth in the germ tubes of the parasitic plants in a manner similar to that in the endogenous tissues of the host plants. The unilateral growth inhibition of the germ tubes may lead to a curvature toward the host plant. Recent research has shown that parasitic plants have evolved a group of α/β-hydroxylases derived from the KAI2 clade of nonparasitic plants, termed the Karrikin-insensitive 2 divergent clade (KAI2d) (Conn et al., Citation2015; Toh et al., Citation2015; Tsuchiya et al., Citation2015). KAI2d family members have been identified for instance in Striga hermonthica and Orobanche minor, where they function as strigolactones receptors for host plant recognition (Conn et al., Citation2015; Toh et al., Citation2015; Tsuchiya et al., Citation2015; Takei et al., Citation2023). A recent study examined the interaction of the STL with members of the O. minor KAI2d clade but showed only a lower affinity of the candidates for the tested STL compared to strigolactones (Takei et al., Citation2023). This suggests that factors other than KAI2d members are required for STL perception in O. minor. In this context, an interesting question for further research would be to study the emergence and functional specificity of the candidates of the KAI2d clade in O. cumana, a host-specific parasite on the STL-exuding sunflower. The results obtained could further contribute to a better understanding of the mechanism of perception and directed growth in response to STL in parasitic plants.
7. Concluding remarks and outlook
Many of the secrets of STL biosynthesis have been unraveled in the past two decades, including the formation of the different lactone rings, the STL backbone conversion and the decoration of the STL backbone. It will be interesting to see what next steps in the biosynthesis of STL will be elucidated in the future. The vast datasets produced by the ever-increasing speed of cutting-edge genome sequencing platforms as well as new methods for the transformation of plants such as improved CRISPR/Cas techniques (Schreiber et al., Citation2023) will contribute to the elucidation of further STL pathways. Elucidation of homologs of STL genes across many Asteraceae lineages and studies of STL biosynthesis enzyme family expansion will help us understand the evolution of STL biosynthesis. A more fine-tuned resolution of STL gene expression and accumulation as well as knock-out of potential STL metabolism regulating genes will help understand the mechanism of their formation in laticifers in the future. The intriguing role of STL as signaling molecules inside plants and their potential role in the search for the so far elusive endogenous Karrikin-insensitive ligand will add a new dimension to our view of this class of specialized metabolites. Understanding the regulation, compartmentalization, and evolution of STL biosynthesis will also help us understand those central aspects for other classes of compounds in plant specialized metabolism.
Supplemental Material
Download MS Excel (37.7 KB)References
- Acquadro, A., Barchi, L., Portis, E., Mangino, G., Valentino, D., Mauromicale, G., and Lanteri, S. 2017. Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation. Sci. Rep. 7:5617. doi:10.1038/s41598-017-05085-7
- Ahern, J. R., and Whitney, K. D. 2014. Sesquiterpene lactone stereochemistry influences herbivore resistance and plant fitness in the field. Ann. Bot. 113:731–740. doi:10.1093/aob/mct297
- Amrehn, E., Aschenbrenner, A. K., Heller, A., and Spring, O. 2016. Localization of sesquiterpene lactone biosynthesis in cells of capitate glandular trichomes of Helianthus annuus (Asteraceae). Protoplasma 253:447–455. doi:10.1007/s00709-015-0823-4
- Andersen, T. B., Martinez-Swatson, K. A., Rasmussen, S. A., Boughton, B. A., Jørgensen, K., Andersen-Ranberg, J., Nyberg, N., Christensen, S. B., and Simonsen, H. T. 2017. Localization and in-vivo characterization of Thapsia garganica CYP76AE2 indicates a role in Thapsigargin biosynthesis. Plant Physiol. 174:56–72. doi:10.1104/pp.16.00055
- Andersen, T. B., Rasmussen, S. A., Christensen, S. B., and Simonsen, H. T. 2019. Biosynthesis of tovarol and other sesquiterpenoids in Thapsia laciniata Rouy. Phytochemistry 157:168–174. doi:10.1016/j.phytochem.2018.10.027
- Andolfi, A., Zermane, N., Cimmino, A., Avolio, F., Boari, A., Vurro, M., and Evidente, A. 2013. Inuloxins A–D, phytotoxic bi-and tri-cyclic sesquiterpene lactones produced by Inula viscosa: potential for broomrapes and field dodder management. Phytochemistry 86:112–120. doi:10.1016/j.phytochem.2012.10.003
- Appezzato-da-Glória, B., Da Costa, F. B., da Silva, V. C., Gobbo-Neto, L., Rehder, V. L. G., and Hayashi, A. H. 2012. Glandular trichomes on aerial and underground organs in Chrysolaena species (Vernonieae – Asteraceae): structure, ultrastructure and chemical composition. Flora Morphol Distribut Funct Ecol Plants 207:878–887. doi:10.1016/j.flora.2012.10.003
- Aschenbrenner, A. K., Amrehn, E., Bechtel, L., and Spring, O. 2015. Trichome differentiation on leaf primordia of Helianthus annuus (Asteraceae): morphology, gene expression and metabolite profile. Planta 241:837–846. doi:10.1007/s00425-014-2223-y
- Aschenbrenner, A. K., Horakh, S., and Spring, O. 2013. Linear glandular trichomes of Helianthus (Asteraceae): morphology, localization, metabolite activity and occurrence. AoB PLANTS 5:plt028–plt028. doi:10.1093/aobpla/plt028
- Badouin, H., Gouzy, J., Grassa, C. J., Murat, F., Staton, S. E., Cottret, L., Lelandais-Brière, C., Owens, G. L., Carrère, S., Mayjonade, B., Legrand, L., Gill, N., Kane, N. C., Bowers, J. E., Hubner, S., Bellec, A., Bérard, A., Bergès, H., Blanchet, N., Boniface, M.-C., Brunel, D., Catrice, O., Chaidir, N., Claudel, C., Donnadieu, C., Faraut, T., Fievet, G., Helmstetter, N., King, M., Knapp, S. J., Lai, Z., Le Paslier, M.-C., Lippi, Y., Lorenzon, L., Mandel, J. R., Marage, G., Marchand, G., Marquand, E., Bret-Mestries, E., Morien, E., Nambeesan, S., Nguyen, T., Pegot-Espagnet, P., Pouilly, N., Raftis, F., Sallet, E., Schiex, T., Thomas, J., Vandecasteele, C., Varès, D., Vear, F., Vautrin, S., Crespi, M., Mangin, B., Burke, J. M., Salse, J., Muños, S., Vincourt, P., Rieseberg, L. H., and Langlade, N. B. 2017. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546:148–152. doi:10.1038/nature22380
- Baek, S., Utomo, J. C., Lee, J. Y., Dalal, K., Yoon, Y. J., and Ro, D.-K. 2021. The yeast platform engineered for synthetic gRNA-landing pads enables multiple gene integrations by a single gRNA/Cas9 system. Metab. Eng. 64:111–121. doi:10.1016/j.ymben.2021.01.011
- Barker, M. S., Li, Z., Kidder, T. I., Reardon, C. R., Lai, Z., Oliveira, L. O., Scascitelli, M., and Rieseberg, L. H. 2016. Most compositae (Asteraceae) are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am. J. Bot. 103:1203–1211. doi:10.3732/ajb.1600113
- Barnes, E. K., Kwon, M., Hodgins, C. L., Qu, Y., Kim, S.-W., Yeung, E. C., and Ro, D.-K. 2021. The promoter sequences of lettuce cis-prenyltransferase and its binding protein specify gene expression in laticifers. Planta 253:51. doi:10.1007/s00425-021-03566-8
- Bellinger, M. R., Datlof, E. M., Selph, K. E., Gallaher, T. J., and Knope, M. L. 2022. A genome for Bidens hawaiensis: a member of a hexaploid Hawaiian plant adaptive radiation. J. Hered. 113:205–214. doi:10.1093/jhered/esab077
- Bennett, M. H., Gallagher, M. D. S., Bestwick, C. S., Rossiter, J. T., and Mansfield, J. W. 1994. The phytoalexin response of lettuce to challenge by Botrytis cinerea, Bremia lactucae and Pseudomonas syringae pv. phaseolicola. Physiol. Mol. Plant Pathol. 44:321–333. doi:10.1016/S0885-5765(05)80046-3
- Berman, P., de Haro, L. A., Jozwiak, A., Panda, S., Pinkas, Z., Dong, Y., Cveticanin, J., Barbole, R., Livne, R., Scherf, T., Shimoni, E., Levin-Zaidman, S., Dezorella, N., Petrovich-Kopitman, E., Meir, S., Rogachev, I., Sonawane, P. D., and Aharoni, A. 2023. Parallel evolution of cannabinoid biosynthesis. Nat. Plants. 9:817–831. doi:10.1038/s41477-023-01402-3
- Bifulco, G., Bruno, I., Paloma, L. G., and Riccio, R. 1993. Edwardsolides A, B and C. New Sesquiterpenoid Lactones from the Mediterranean Octocoral Maasella edwardsi. Natural Product Letters 3:167–171. doi:10.1080/10575639308043857
- Bird, D. A., Franceschi, V. R., and Facchini, P. J. 2003. A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell. 15:2626–2635. doi:10.1105/tpc.015396
- Bogdanović, M., Cankar, K., Todorović, S., Dragicević, M., Simonović, A., van Houwelingen, A., Schijlen, E., Schipper, B., Gagneul, D., Hendriks, T., Quillet, M.-C., Bouwmeester, H., Bosch, D., and Beekwilder, J. 2019. Tissue specific expression and genomic organization of bitter sesquiterpene lactone biosynthesis in Cichorium intybus L. (Asteraceae). Ind. Crops Prod. 129:253–260. doi:10.1016/j.indcrop.2018.12.011
- Bouwmeester, H. J., Kodde, J., Verstappen, F. W. a., Altug, I. G., de Kraker, J.-W., and Wallaart, T. E. 2002. Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol. 129:134–144. doi:10.1104/pp.001024
- Cankar, K., Bundock, P., Sevenier, R., Häkkinen, S. T., Hakkert, J. C., Beekwilder, J., van der Meer, I. M., de Both, M., and Bosch, D. 2021. Inactivation of the germacrene A synthase genes by CRISPR/Cas9 eliminates the biosynthesis of sesquiterpene lactones in Cichorium intybus L. Plant Biotechnol. J. 19:2442–2453. doi:10.1111/pbi.13670
- Cankar, K., Hakkert, J. C., Sevenier, R., Campo, E., Schipper, B., Papastolopoulou, C., Vahabi, K., Tissier, A., Bundock, P., and Bosch, D. 2022. CRISPR/Cas9 targeted inactivation of the kauniolide synthase in chicory results in accumulation of costunolide and its conjugates in taproots. Front. Plant Sci. 13:940003. doi:10.3389/fpls.2022.940003
- Cankar, K., Hakkert, J. C., Sevenier, R., Papastolopoulou, C., Schipper, B., Baixinho, J. P., Fernández, N., Matos, M. S., Serra, A. T., Santos, C. N., Vahabi, K., Tissier, A., Bundock, P., and Bosch, D. 2023. Lactucin synthase inactivation boosts the accumulation of anti-inflammatory 8-deoxylactucin and its derivatives in chicory (Cichorium intybus L.). J. Agric. Food Chem. 71:6061–6072. doi:10.1021/acs.jafc.2c08959
- Cankar, K., Houwelingen, A. V., Bosch, D., Sonke, T., Bouwmeester, H., and Beekwilder, J. 2011. A chicory cytochrome P450 mono-oxygenase CYP71AV8 for the oxidation of (+)-valencene. FEBS Lett. 585:178–182. doi:10.1016/j.febslet.2010.11.040
- Catalán, C. A. N., Borkosky, S. A., and Joseph-Nathan, P. 1996. The secondary metabolite chemistry of the subtribe Gochnatiinae (tribe Mutisieae, family Compositae). Biochem. Syst. Ecol. 24:659–718. doi:10.1016/S0305-1978(96)00063-4
- Catania, T. M., Branigan, C. A., Stawniak, N., Hodson, J., Harvey, D., Larson, T. R., Czechowski, T., and Graham, I. A. 2018. Silencing amorpha-4,11-diene synthase genes in Artemisia annua Leads to FPP Accumulation. Front. Plant Sci. 9:547. doi:10.3389/fpls.2018.00547
- Cavallito, C. J., and Haskell, T. H. 1945. The mechanism of action of antibiotics. The reaction of unsaturated lactones with cysteine and related compounds. J. Am. Chem. Soc. 67:1991–1994. doi:10.1021/ja01227a037
- Chang, M. C. Y., Eachus, R. a., Trieu, W., Ro, D.-K., and Keasling, J. D. 2007. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 3:274–277. doi:10.1038/nchembio875
- Chen, H., Guo, M., Dong, S., Wu, X., Zhang, G., He, L., Jiao, Y., Chen, S., Li, L., and Luo, H. 2023. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 4:100516. doi:10.1016/j.xplc.2023.100516
- Conn, C. E., Bythell-Douglas, R., Neumann, D., Yoshida, S., Whittington, B., Westwood, J. H., Shirasu, K., Bond, C. S., Dyer, K. A., and Nelson, D. C. 2015. PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349:540–543. doi:10.1126/science.aab1140
- Czechowski, T., Larson, T. R., Catania, T. M., Harvey, D., Brown, G. D., and Graham, I. A. 2016. Artemisia annua mutant impaired in artemisinin synthesis demonstrates importance of nonenzymatic conversion in terpenoid metabolism. Proc. Natl. Acad. Sci. U S A. 113:15150–15155. doi:10.1073/pnas.1611567113
- De Bruyn, C., Ruttink, T., Lacchini, E., Rombauts, S., Haegeman, A., De Keyser, E., Van Poucke, C., Desmet, S., Jacobs, T. B., Eeckhaut, T., Goossens, A., and Van Laere, K. 2023. Identification and characterization of CYP71 subclade cytochrome P450 enzymes involved in the biosynthesis of bitterness compounds in Cichorium intybus. Front. Plant Sci. 14:1200253. doi:10.3389/fpls.2023.1200253
- Degenhardt, J., Köllner, T. G., and Gershenzon, J. 2009. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70:1621–1637. doi:10.1016/j.phytochem.2009.07.030
- Desgagné-Penix, I., and Facchini, P. J. 2012. Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 72:331–344. doi:10.1111/j.1365-313X.2012.05084.x
- Duke, S. O., and Paul, R. N. 1993. Development and fine structure of the Glandular Trichomes of Artemisia annua L. International Journal of Plant Sciences 154:107–118. doi:10.1086/297096
- Duke, M. V., Paul, R. N., Elsohly, H. N., Sturtz, G., and Duke, S. O. 1994. Localization of Artemisinin and Artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. Int. J. Plant Sci. 155:365–372. doi:10.1086/297173
- Eljounaidi, K., Cankar, K., Comino, C., Moglia, A., Hehn, A., Bourgaud, F. d. r., Bouwmeester, H., Menin, B., Lanteri, S., and Beekwilder, J. 2014. Cytochrome P450s from Cynara cardunculus L. CYP71AV9 and CYP71BL5, catalyze distinct hydroxylations in the sesquiterpene lactone biosynthetic pathway. Plant Sci. 223:59–68. doi:10.1016/j.plantsci.2014.03.007
- Fahn, A. 2000. Structure and function of secretory cells. Adv. Bot. Res. 31:37–66. doi:10.1016/S0065-2296(00)31006-0
- Fan, W., Wang, S., Wang, H., Wang, A., Jiang, F., Liu, H., Zhao, H., Xu, D., and Zhang, Y. 2022. The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae palaeo-polyploidization history and plant inulin production. Mol. Ecol. Resour. 22:3124–3140. doi:10.1111/1755-0998.13675
- Fischer, N. H., Olivier, E. J., and Fischer, H. D. 1979. The Biogenesis and Chemistry of Sesquiterpene Lactones BT - Fortschritte Der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products; Fischer H. D., Fischer N. H., Franck R. W., Eds. Vienna, Springer Vienna, pp. 47–320.
- Frey, M. 2020. Traps and pitfalls-unspecific reactions in metabolic engineering of Sesquiterpenoid pathways. Molecules 25:1935. doi:10.3390/molecules25081935
- Frey, M., Klaiber, I., Conrad, J., and Spring, O. 2020. CYP71BL9, the missing link in costunolide synthesis of sunflower. Phytochemistry 177:112430. doi:10.1016/j.phytochem.2020.112430
- Frey, M., Klaiber, I., Conrad, J., Bersch, A., Pateraki, I., Ro, D.-K., and Spring, O. 2019. Characterization of CYP71AX36 from Sunflower (Helianthus annuus L., Asteraceae). Sci. Rep. 9:14295–14295. doi:10.1038/s41598-019-50520-6
- Frey, M., Schmauder, K., Pateraki, I., and Spring, O. 2018. Biosynthesis of Eupatolide-a metabolic route for Sesquiterpene lactone formation involving the P450 enzyme CYP71DD6. ACS Chem. Biol. 13:1536–1543. doi:10.1021/acschembio.8b00126
- Fuentes, P., Zhou, F., Erban, A., Karcher, D., Kopka, J., and Bock, R. 2016. A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife 5:e13664.
- Ghantous, A., Gali-Muhtasib, H., Vuorela, H., Saliba, N. A., and Darwiche, N. 2010. What made sesquiterpene lactones reach cancer clinical trials? Drug Discov. Today. 15:668–678. doi:10.1016/j.drudis.2010.06.002
- Göpfert, J., Bülow, A. K., and Spring, O. 2010. Identification and functional characterization of a new sunflower germacrene A synthase (HaGAS3). Nat. Prod. Commun. 5:1934578X1000500. doi:10.1177/1934578X1000500507
- Göpfert, J., Conrad, J., and Spring, O. 2006. 5-deoxynevadensin, a novel flavone in sunflower and aspects of biosynthesis during trichome development. Nat. Prod. Commun. 1:1934578X0600101. doi:10.1177/1934578X0600101104
- Göpfert, J. C., Heil, N., Conrad, J., and Spring, O. 2005. Cytological development and sesquiterpene lactone secretion in capitate glandular trichomes of sunflower. Plant Biol. (Stuttg) 7:148–155. doi:10.1055/s-2005-837575
- Göpfert, J. C., Macnevin, G., Ro, D.-K., and Spring, O. 2009. Identification, functional characterization and developmental regulation of sesquiterpene synthases from sunflower capitate glandular trichomes. BMC Plant Biol. 9:86–86. doi:10.1186/1471-2229-9-86
- Gou, J., Hao, F., Huang, C., Kwon, M., Chen, F., Li, C., Liu, C., Ro, D.-K., Tang, H., and Zhang, Y. 2017. Discovery of a non-stereoselective cytochrome P450 catalyzing either 8α- or 8β-hydroxylation of germacrene A acid from the Chinese medicinal plant, Inula hupehensis. Plant J. 93:92–106. doi:10.1111/tpj.13760
- Greinwald, A., Hartmann, M., Heilmann, J., Heinrich, M., Luick, R., and Reif, A. 2022. Soil and vegetation drive Sesquiterpene lactone content and profile in Arnica montana L. Flower heads from Apuseni-mountains, Romania. Front. Plant Sci. 13:813939. doi:10.3389/fpls.2022.813939
- Gutiérrez, D. G., and Luna, M. L. 2013. A comparative study of latex-producing tissues in genera of Liabeae (Asteraceae). Flora Morphol Distribut Function Ecol Plants 208:33–44. doi:10.1016/j.flora.2012.11.001
- Hagel, J. M., Yeung, E. C., and Facchini, P. J. 2008. Got milk? The secret life of laticifers. Trends Plant Sci. 13:631–639. doi:10.1016/j.tplants.2008.09.005
- Han, J., Wang, H., Kanagarajan, S., Hao, M., Lundgren, A., and Brodelius, P. E. 2016. Promoting Artemisinin biosynthesis in Artemisia annua plants by substrate channeling. Mol. Plant. 9:946–948. doi:10.1016/j.molp.2016.03.004
- Hansen, C. C., Nelson, D. R., Møller, B. L., and Werck-Reichhart, D. 2021. Plant cytochrome P450 plasticity and evolution. Mol. Plant. 14:1772. doi:10.1016/j.molp.2021.09.013
- Hassani, D., Taheri, A., Fu, X., Qin, W., Hang, L., Ma, Y., and Tang, K. 2023. Elevation of artemisinin content by co-transformation of artemisinin biosynthetic pathway genes and trichome-specific transcription factors in Artemisia annua. Front. Plant Sci. 14:1118082. doi:10.3389/fpls.2023.1118082
- He, X., Sun, Y., and Zhu, R.-L. 2013. The oil bodies of liverworts: unique and important organelles in land plants. Crit. Rev. Plant Sci. 32:293–302. doi:10.1080/07352689.2013.765765
- Henquet, M. G. L., Prota, N., van der Hooft, J. J. J., Varbanova-Herde, M., Hulzink, R. J. M., de Vos, M., Prins, M., de Both, M. T. J., Franssen, M. C. R., Bouwmeester, H., and Jongsma, M. 2017. Identification of a drimenol synthase and drimenol oxidase from Persicaria hydropiper, involved in the biosynthesis of insect deterrent drimanes. Plant J. 90:1052–1063. doi:10.1111/tpj.13527
- Hingsamer, M., Kulmer, V., de Roode, M., and Kernitzkyi, M. 2022. Environmental and socio-economic impacts of new plant breeding technologies: a case study of root chicory for inulin production. Front. Genome Ed. 4:919392. doi:10.3389/fgeed.2022.919392
- Huang, S. H., Duke, R. K., Chebib, M., Sasaki, K., Wada, K., and Johnston, G. A. R. 2003. Bilobalide, a sesquiterpene trilactone from Ginkgo biloba, is an antagonist at recombinant α1β2γ2L GABAA receptors. Eur. J. Pharmacol. 464:1–8. doi:10.1016/s0014-2999(03)01344-x
- Huber, M., Epping, J., Schulze Gronover, C., Fricke, J., Aziz, Z., Brillatz, T., Swyers, M., Köllner, T. G., Vogel, H., Hammerbacher, A., Triebwasser-Freese, D., Robert, C. A. M., Verhoeven, K., Preite, V., Gershenzon, J., and Erb, M. 2016. A latex metabolite benefits plant fitness under root herbivore attack. PLoS Biol. 14:e1002332. doi:10.1371/journal.pbio.1002332
- Ikezawa, N., Göpfert, J. C., Nguyen, D. T., Kim, S. U., O'Maille, P. E., Spring, O., and Ro, D. K. 2011. Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. J. Biol. Chem. 286:21601–21611. doi:10.1074/jbc.M110.216804
- Iorizzo, M., Ellison, S., Senalik, D., Zeng, P., Satapoomin, P., Huang, J., Bowman, M., Iovene, M., Sanseverino, W., Cavagnaro, P., Yildiz, M., Macko-Podgórni, A., Moranska, E., Grzebelus, E., Grzebelus, D., Ashrafi, H., Zheng, Z., Cheng, S., Spooner, D., Van Deynze, A., and Simon, P. 2016. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48:657–666. doi:10.1038/ng.3565
- Joel, D. M., Chaudhuri, S. K., Plakhine, D., Ziadna, H., and Steffens, J. C. 2011. Dehydrocostus lactone is exuded from sunflower roots and stimulates germination of the root parasite Orobanche cumana. Phytochemistry 72:624–634. doi:10.1016/j.phytochem.2011.01.037
- Kalsi, P. S., Kaur, P., and Chhabra, B. R. 1979. Plant growth activity of epoxides from dehydrocostus lactone. Phytochemistry 18:1877–1878. doi:10.1016/0031-9422(79)83076-9
- King, B. C., Vavitsas, K., Ikram, N. K. B. K., Schrøder, J., Scharff, L. B., Bassard, J.-É., Hamberger, B., Jensen, P. E., and Simonsen, H. T. 2016. In vivo assembly of DNA-fragments in the moss, Physcomitrella patens. Sci. Rep. 6:25030–25030. doi:10.1038/srep25030
- Knoche, H., Ourisson, G., Perold, G. W., Foussereau, J., and Maleville, J. 1969. Allergenic component of a liverwort: sesquiterpene lactone. Science 166:239–240. doi:10.1126/science.166.3902.239
- Krupp, A., Bertsch, B., and Spring, O. 2021. Costunolide influences germ tube orientation in sunflower broomrape – a first step toward understanding chemotropism. Front. Plant Sci. 12:699068. doi:10.3389/fpls.2021.699068
- Kupchan, S. M., Fessler, D. C., Eakin, M. A., and Giacobbe, T. J. 1970. Reactions of alpha methylene lactone tumor inhibitors with model biological nucelophiles. Science 168:376–378. doi:10.1126/science.168.3929.376
- Kwon, M., Hodgins, C. L., Haslam, T. M., Roth, S. A., Nguyen, T.-D., Yeung, E. C., and Ro, D.-K. 2022. Germacrene a synthases for sesquiterpene lactone biosynthesis are expressed in vascular parenchyma cells neighboring laticifers in lettuce. Plants 11:1192. doi:10.3390/plants11091192
- Kwon, M., Hodgins, C. L., Salama, E. M., Dias, K. R., Parikh, A., Mackey, A. V., Catenza, K. F., Vederas, J. C., and Ro, D. K. 2023. New insights into natural rubber biosynthesis from rubber‐deficient lettuce mutants expressing goldenrod or guayule cis‐prenyltransferase. New Phytol. 239:1098–1111. doi:10.1111/nph.18994
- Lange, B. M. 2015. The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu. Rev. Plant Biol. 66:139–159. doi:10.1146/annurev-arplant-043014-114639
- Liao, B., Shen, X., Xiang, L., Guo, S., Chen, S., Meng, Y., Liang, Y., Ding, D., Bai, J., Zhang, D., Czechowski, T., Li, Y., Yao, H., Ma, T., Howard, C., Sun, C., Liu, H., Liu, J., Pei, J., Gao, J., Wang, J., Qiu, X., Huang, Z., Li, H., Yuan, L., Wei, J., Graham, I., Xu, J., Zhang, B., and Chen, S. 2022. Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield. Mol. Plant. 15:1310–1328. doi:10.1016/j.molp.2022.05.013
- Liu, Q., Beyraghdar Kashkooli, A., Manzano, D., Pateraki, I., Richard, L., Kolkman, P., Lucas, M. F., Guallar, V., de Vos, R. C. H., Franssen, M. C. R., van der Krol, A., and Bouwmeester, H. 2018. Kauniolide synthase is a P450 with unusual hydroxylation and cyclization-elimination activity. Nat. Commun. 9:4657–4657. doi:10.1038/s41467-018-06565-8
- Liu, Q., Majdi, M., Cankar, K., Goedbloed, M., Charnikhova, T., Verstappen, F. W. A., de Vos, R. C. H., Beekwilder, J., van der Krol, S., and Bouwmeester, H. J. 2011. Reconstitution of the costunolide biosynthetic pathway in yeast and Nicotiana benthamiana. PLoS One. 6:e23255. doi:10.1371/journal.pone.0023255
- Liu, Q., Manzano, D., Tanić, N., Pesic, M., Bankovic, J., Pateraki, I., Ricard, L., Ferrer, A., de Vos, R., de Krol, S. v., and Bouwmeester, H. 2014. Elucidation and in planta reconstitution of the parthenolide biosynthetic pathway. Metab. Eng. 23:145–153. doi:10.1016/j.ymben.2014.03.005
- Liu, B., Yan, J., Li, W., Yin, L., Li, P., Yu, H., Xing, L., Cai, M., Wang, H., Zhao, M., Zheng, J., Sun, F., Wang, Z., Jiang, Z., Ou, Q., Li, S., Qu, L., Zhang, Q., Zheng, Y., Qiao, X., Xi, Y., Zhang, Y., Jiang, F., Huang, C., Liu, C., Ren, Y., Wang, S., Liu, H., Guo, J., Wang, H., Dong, H., Peng, C., Qian, W., Fan, W., and Wan, F. 2020. Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 11:340. doi:10.1038/s41467-019-13926-4
- Lu, X., Zhang, L., Zhang, F., Jiang, W., Shen, Q., Zhang, L., Lv, Z., Wang, G., and Tang, K. 2013. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198:1191–1202. doi:10.1111/nph.12207
- Lv, Z., Wang, S., Zhang, F., Chen, L., Hao, X., Pan, Q., Fu, X., Li, L., Sun, X., and Tang, K. 2016. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of Artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 57:1961–1971. doi:10.1093/pcp/pcw118
- Macías, F. A., Galindo, J. C. G., Castellano, D., and Velasco, R. F. 1999. Sesquiterpene lactones with potential use as natural herbicide models (I): trans,trans- germacranolides. J. Agric. Food Chem. 47:4407–4414. doi:10.1021/jf9903612
- Macías, F. A., Torres, A., Molinllo, J. M.G., Varela, R. M., and Castellano, D. 1996. Potential allelopathic sesquiterpene lactones from sunflower leaves. Phytochemistry 43:1205–1215. doi:10.1016/S0031-9422(96)00392-5
- Maes, L., Van Nieuwerburgh, F. C. W., Zhang, Y., Reed, D. W., Pollier, J., Vande Casteele, S. R. F., Inzé, D., Covello, P. S., Deforce, D. L. D., and Goossens, A. 2011. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189:176–189. doi:10.1111/j.1469-8137.2010.03466.x
- Magnusson, G., Thorén, S., Lindman, B., Merényi, G., van der Hoeven, M. G., and Swahn, C.-G. 1973. Fungal extractives. III. Two sesquiterpene lactones from Lactarius. Acta Chem. Scand. 27:1573–1578. doi:10.3891/acta.chem.scand.27-1573
- Majdi, M., Abdollahi, M. R., and Maroufi, A. 2015. Parthenolide accumulation and expression of genes related to parthenolide biosynthesis affected by exogenous application of methyl jasmonate and salicylic acid in Tanacetum parthenium. Plant Cell Rep. 34:1909–1918. doi:10.1007/s00299-015-1837-2
- Majdi, M., Ashengroph, M., and Abdollahi, M. R. 2016. Sesquiterpene lactone engineering in microbial and plant platforms: parthenolide and artemisinin as case studies. Appl. Microbiol. Biotechnol. 100:1041–1059. doi:10.1007/s00253-015-7128-6
- Majdi, M., Charnikhova, T., and Bouwmeester, H. 2013. Genetical, developmental and spatial factors influencing parthenolide and its precursor costunolide in feverfew (Tanacetum parthenium L. Schulz Bip.). Ind. Crops Prod. 47:270–276. doi:10.1016/j.indcrop.2013.03.021
- Mandel, J. R., Dikow, R. B., Siniscalchi, C. M., Thapa, R., Watson, L. E., and Funk, V. A. 2019. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc. Natl. Acad. Sci. U S A. 116:14083–14088. doi:10.1073/pnas.1903871116
- Martínez-Quezada, D. M., Rivera, P., Rojas-Leal, A., Villaseñor, J. L., and Terrazas, T. 2023. Leaf secretory structures in asteraceae: a synthesis of their diversity and evolution. Bot. Rev. 89:59–90. doi:10.1007/s12229-022-09276-4
- McEvoy, S. L., Lustenhouwer, N., Melen, M. K., Nguyen, O., Marimuthu, M. P. A., Chumchim, N., Beraut, E., Parker, I. M., and Meyer, R. S. 2023. Chromosome-level reference genome of stinkwort, Dittrichia graveolens (L.) Greuter: a resource for studies on invasion, range expansion, and evolutionary adaptation under global change. J. Hered. 114:561–569. doi:10.1093/jhered/esad033
- Mercke, P., Bengtsson, M., Bouwmeester, H. J., Posthumus, M. A., and Brodelius, P. E. 2000. Molecular cloning, expression, and characterization of Amorpha-4,11-diene synthase, a key enzyme of Artemisinin biosynthesis in Artemisia annua L. Arch. Biochem. Biophys. 381:173–180. doi:10.1006/abbi.2000.1962
- Michael, A. 1887. Ueber die Addition von Natriumacetessig- und Natriummalonsaureathern zu den Aethern ungesattigter Sauren. J. Prakt. Chem. 35:349–356. doi:10.1002/prac.18870350136
- Morimoto, H., Sanno, Y., and Oshio, H. 1966. Chemical studies on heliangine. Tetrahedron 22:3173–3179. doi:10.1016/S0040-4020(01)82296-1
- Nguyen, T. D., Faraldos, J. A., Vardakou, M., Salmon, M., O'Maille, P. E., and Ro, D. K. 2016. Discovery of germacrene A synthases in Barnadesia spinosa: the first committed step in sesquiterpene lactone biosynthesis in the basal member of the Asteraceae. Biochem. Biophys. Res. Commun. 479:622–627. doi:10.1016/j.bbrc.2016.09.165
- Nguyen, D. T., Göpfert, J. C., Ikezawa, N., MacNevin, G., Kathiresan, M., Conrad, J., Spring, O., and Ro, D. K. 2010. Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J. Biol. Chem. 285:16588–16598. doi:10.1074/jbc.M110.111757
- Nguyen, T.-D., Kwon, M., Kim, S.-U., Fischer, C., and Ro, D.-K. 2019. Catalytic plasticity of germacrene A oxidase underlies sesquiterpene lactone diversification. Plant Physiol. 181:945–960. doi:10.1104/pp.19.00629
- Nützmann, H.-W., and Osbourn, A. 2014. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26:91–99. doi:10.1016/j.copbio.2013.10.009
- Olofsson, L., Lundgren, A., and Brodelius, P. E. 2012. Trichome isolation with and without fixation using laser microdissection and pressure catapulting followed by RNA amplification: expression of genes of terpene metabolism in apical and sub-apical trichome cells of Artemisia annua L. Plant Sci. 183:9–13. doi:10.1016/j.plantsci.2011.10.019
- Olsson, M. E., Olofsson, L. M., Lindahl, A.-L., Lundgren, A., Brodelius, M., and Brodelius, P. E. 2009. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70:1123–1128. doi:10.1016/j.phytochem.2009.07.009
- Onoyovwe, A., Hagel, J. M., Chen, X., Khan, M. F., Schriemer, D. C., and Facchini, P. J. 2013. Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers. Plant Cell. 25:4110–4122. doi:10.1105/tpc.113.115113
- Ozber, N., Carr, S. C., Morris, J. S., Liang, S., Watkins, J. L., Caldo, K. M., Hagel, J. M., Ng, K. K. S., and Facchini, P. J. 2022. Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation. Nat. Commun. 13:6768. doi:10.1038/s41467-022-34313-6
- Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., Leavell, M. D., Tai, a., Main, a., Eng, D., Polichuk, D. R., Teoh, K. H., Reed, D. W., Treynor, T., Lenihan, J., Fleck, M., Bajad, S., Dang, G., Dengrove, D., Diola, D., Dorin, G., Ellens, K. W., Fickes, S., Galazzo, J., Gaucher, S. P., Geistlinger, T., Henry, R., Hepp, M., Horning, T., Iqbal, T., Jiang, H., Kizer, L., Lieu, B., Melis, D., Moss, N., Regentin, R., Secrest, S., Tsuruta, H., Vazquez, R., Westblade, L. F., Xu, L., Yu, M., Zhang, Y., Zhao, L., Lievense, J., Covello, P. S., Keasling, J. D., Reiling, K. K., Renninger, N. S., and Newman, J. D. 2013. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. doi:10.1038/nature12051
- Padilla-González, G. F., Amrehn, E., Frey, M., Gómez-Zeledón, J., Kaa, A., Da Costa, F. B., and Spring, O. 2020. Metabolomic and gene expression studies reveal the diversity, distribution and spatial regulation of the specialized metabolism of yacón (Smallanthus sonchifolius, asteraceae). Int J Mol Sci 21:4555. doi:10.3390/ijms21124555
- Padilla-González, G. F., Frey, M., Gómez-Zeledón, J., Da Costa, F. B., and Spring, O. 2019. Metabolomic and gene expression approaches reveal the developmental and environmental regulation of the secondary metabolism of yacón (Smallanthus sonchifolius, Asteraceae). Sci. Rep. 9:13178. doi:10.1038/s41598-019-49246-2
- Padilla-Gonzalez, G. F., dos Santos, F. A., and Da Costa, F. B. 2016. Sesquiterpene lactones: more than protective plant compounds with high toxicity. Crit. Rev. Plant Sci. 35:18–37. doi:10.1080/07352689.2016.1145956
- Peng, Y., Lai, Z., Lane, T., Nageswara-Rao, M., Okada, M., Jasieniuk, M., O'Geen, H., Kim, R. W., Sammons, R. D., Rieseberg, L. H., and Stewart, C. N. Jr. 2014. De Novo genome assembly of the economically important weed horseweed using integrated data from multiple sequencing platforms. Plant Physiol. 166:1241–1254. doi:10.1104/pp.114.247668
- Perry, N. B., Burgess, E. J., Rodríguez Guitián, M. A., Romero Franco, R., López Mosquera, E., Smallfield, B. M., Joyce, N. I., and Littlejohn, R. P. 2009. Sesquiterpene lactones in Arnica montana: helenalin and dihydrohelenalin chemotypes in Spain. Planta Med. 75:660–666. doi:10.1055/s-0029-1185362
- Pickel, B., Drew, D. P., Manczak, T., Weitzel, C., Simonsen, H. T., and Ro, D.-K. 2012. Identification and characterization of a kunzeaol synthase from Thapsia garganica: implications for the biosynthesis of the pharmaceutical thapsigargin. Biochem. J. 448:261–271. doi:10.1042/BJ20120654
- Picman, A. K. 1986. Biological activities of Sesquiterpene lactones. Biochem. Syst. Ecol. 14:255–281. doi:10.1016/0305-1978(86)90101-8
- Pu, G.-B., Ma, D.-M., Chen, J.-L., Ma, L.-Q., Wang, H., Li, G.-F., Ye, H.-C., and Liu, B.-Y. 2009. Salicylic acid activates artemisinin biosynthesis in Artemisia annua L. Plant Cell Rep. 28:1127–1135. doi:10.1007/s00299-009-0713-3
- Qu, Y., Chakrabarty, R., Tran, H. T., Kwon, E.-J G., Kwon, M., Nguyen, T.-D., and Ro, D.-K. 2015. A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 290:1898–1914. doi:10.1074/jbc.M114.616920
- Rahimi, M., and Bouwmeester, H. 2021. Are sesquiterpene lactones the elusive KARRIKIN-INSENSITIVE2 ligand? Planta 253:54–54. doi:10.1007/s00425-021-03571-x
- Ramirez, A. M., Saillard, N., Yang, T., Franssen, M. C. R., Bouwmeester, H. J., and Jongsma, M. A. 2013. Biosynthesis of Sesquiterpene lactones in Pyrethrum (Tanacetum cinerariifolium). PLoS One. 8:e65030. doi:10.1371/journal.pone.0065030
- Raupp, F. M., and Spring, O. 2013. New sesquiterpene lactones from sunflower root exudate as germination stimulants for Orobanche cumana. J. Agric. Food Chem. 61:10481–10487. doi:10.1021/jf402392e
- Ro, D. K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., Ho, K. A., Eachus, R. A., Ham, T. S., Kirby, J., Chang, M. C., Withers, S. T., Shiba, Y., Sarpong, R., and Keasling, J. D. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943. doi:10.1038/nature04640
- Roberfroid, M. B. 2007. Inulin-type fructans: functional food ingredients. J. Nutr. 137:2493S–2502S. doi:10.1093/jn/137.11.2493S
- Rodriguez, E., Towers, G. H. N. H. N., and Mitchell, J. C. C. 1976. Biological activities of sesquiterpene lactones. Phytochemistry 15:1573–1580. doi:10.1016/S0031-9422(00)97430-2
- Rozpądek, P., Wężowicz, K., Stojakowska, A., Malarz, J., Surówka, E., Sobczyk, Ł., Anielska, T., Ważny, R., Miszalski, Z., and Turnau, K. 2014. Mycorrhizal fungi modulate phytochemical production and antioxidant activity of Cichorium intybus L. (Asteraceae) under metal toxicity. Chemosphere 112:217–224. doi:10.1016/j.chemosphere.2014.04.023
- Scavo, A., Rial, C., Molinillo, J. M. G., Varela, R. M., Mauromicale, G., and Macı As, F. A. 2020. Effect of shading on the sesquiterpene lactone content and phytotoxicity of cultivated cardoon leaf extracts. J. Agric. Food Chem. 68:11946–11953. doi:10.1021/acs.jafc.0c03527
- Schilling, E. E. 1983. Flavonoids of helianthus series Angustifolii. Biochem. Syst. Ecol. 11:341–344. doi:10.1016/0305-1978(83)90034-0
- Schmidt, T. J. 1999. Toxic activities of sesquiterpene lactones: structural and biochemical aspects. Curr. Org. Chem. 3:577–608.
- Schmidt, T. J. 2006. Structure-activity relationships of sesquiterpene lactones. Stud. Nat. Prod. Chem. 309–392.
- Schreiber, T., Prange, A., Schäfer, P., Iwen, T., Grützner, R., Marillonnet, S., and Tissier, A. 2023. Efficient scar-free knock-ins of several kilobases by engineered CRISPR/Cas endonucleases. bioRxiv 2023.2011.2024.568561.
- Seaman, F. C. 1982. Sesquiterpene lactones as taxonomic characters in the asteraceae. Bot. Rev. 48:121–594. doi:10.1007/BF02919190
- Seemann, A., Wallner, T., Poschlod, P., and Heilmann, J. 2010. Variation of Sesquiterpene lactone contents in different Arnica montana populations: influence of ecological parameters. Planta Med. 76:837–842. doi:10.1055/s-0029-1240797
- Sessa, R. A., Bennett, M., Lewis, M., Mansfield, J., and Beale, M. 2000. Metabolite profiling of sesquiterpene lactones from Lactuca species. Major latex components are novel oxalate and sulfate conjugates of lactucin and its derivatives. J. Biol. Chem. 275:26877–26884. doi:10.1016/S0021-9258(19)61456-0
- Shen, F., He, H., Huang, X., Deng, Y., and Yang, X. 2023a. Insights into the convergent evolution of fructan biosynthesis in angiosperms from the highly characteristic chicory genome. New Phytol. 238:1245–1262. doi:10.1111/nph.18796
- Shen, F., Qin, Y., Wang, R., Huang, X., Wang, Y., Gao, T., He, J., Zhou, Y., Jiao, Y., Wei, J., Li, L., and Yang, X. 2023b. Comparative genomics reveals a unique nitrogen-carbon balance system in Asteraceae. Nat. Commun. 14:4334–4334. doi:10.1038/s41467-023-40002-9
- Shen, Q., Zhang, L., Liao, Z., Wang, S., Yan, T., Shi, P., Liu, M., Fu, X., Pan, Q., Wang, Y., Lv, Z., Lu, X., Zhang, F., Jiang, W., Ma, Y., Chen, M., Hao, X., Li, L., Tang, Y., Lv, G., Zhou, Y., Sun, X., Brodelius, P. E., Rose, J. K. C., and Tang, K. 2018. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and Artemisinin biosynthesis. Mol. Plant. 11:776–788. doi:10.1016/j.molp.2018.03.015
- Shibaoka, H. 1961. Studies on the mechanism of growth inhibiting effect of light. Plant Cell Physiol 2:175–197.
- Singh, I. P., Talwar, K. K., Arora, J. K., Chhabra, B. R., and Kalsi, P. S. 1992. A biologically active guaianolide from Saussurea lappa. Phytochemistry 31:2529–2531. doi:10.1016/0031-9422(92)83317-R
- Spring, O. 2000. Chemotaxonomy based on metabolites from glandular trichomes. Adv Bot Res 31:153–174.
- Spring, O. 2021. Sesquiterpene lactones in sunflower oil. LWT 142:111047–111047. doi:10.1016/j.lwt.2021.111047
- Spring, O., Albert, K., and Gradmann, W. 1981. Annuithrin, a new biologically active germacranolide from Helianthus annuus. Phytochemistry 20:1883–1885. doi:10.1016/0031-9422(81)84027-7
- Spring, O., and Bienert, U. 1987. Capitate glandular hairs from sunflower leaves: development, distribution and sesquiterpene lactone content. J. Plant Physiol. 130:441–448. doi:10.1016/S0176-1617(87)80209-2
- Spring, O., Bienert, U., and Klemt, V. 1987. Sesquiterpene lactones in glandular trichomes of sunflower leaves. J. Plant Physiol. 130:433–439. doi:10.1016/S0176-1617(87)80208-0
- Spring, O., and Hager, A. 1982. Inhibition of elongation growth by two sesquiterpene lactones isolated from Helianthus annuus L. - possible molecular mechanism. Planta 156:433–440. doi:10.1007/BF00393314
- Spring, O., Priester, T., and Hager, A. 1986. Light-induced accumulation of sesquiterpene lactones in sunflower seedlings. J. Plant Physiol. 123:79–89. doi:10.1016/S0176-1617(86)80068-2
- Spring, O., and Schilling, E. E. 1991. The sesquiterpene lactone chemistry of Helianthus sect. Atrorubentes (Asteraceae: Heliantheae). Biochem. Syst. Ecol. 19:59–79. doi:10.1016/0305-1978(91)90114-F
- Spring, O., Schmauder, K., Lackus, N. D., Schreiner, J., Meier, C., Wellhausen, J., Smith, L. V., and Frey, M. 2020. Spatial and developmental synthesis of endogenous sesquiterpene lactones supports function in growth regulation of sunflower. Planta 252:2. doi:10.1007/s00425-020-03409-y
- Spring, O., Zitterell-Haid, B., Bierner, M. W., and Mabry, T. J. 1994. Chemistry of glandular trichomes in Hymenoxys and related genera. Biochem. Syst. Ecol. 22:171–195. doi:10.1016/0305-1978(94)90007-8
- Stojanović, M., Savić, S., Delcourt, A., Hilbert, J.-L., Hance, P., Dragišić Maksimović, J., and Maksimović, V. 2023. Phenolics and Sesquiterpene lactones profile of red and green lettuce: combined effect of cultivar, microbiological fertiliser, and season. Plants (Basel) 12:2616. doi:10.3390/plants12142616
- Sy, L. K., and Brown, G. D. 2002. The mechanism of the spontaneous autoxidation of dihydroartemisinic acid. Tetrahedron 58:897–908. doi:10.1016/S0040-4020(01)01193-0
- Takahashi, S., Yeo, Y. S., Zhao, Y., O'Maille, P. E., Greenhagen, B. T., Noel, J. P., Coates, R. M., and Chappell, J. 2007. Functional characterization of premnaspirodiene oxygenase, a cytochrome P450 catalyzing regio- and stereo-specific hydroxylations of diverse sesquiterpene substrates. J. Biol. Chem. 282:31744–31754. doi:10.1074/jbc.M703378200
- Takei, S., Uchiyama, Y., Bürger, M., Suzuki, T., Okabe, S., Chory, J., and Seto, Y. 2023. A divergent Clade KAI2 protein in the root parasitic plant Orobanche minor is a highly sensitive strigolactone receptor and is involved in the perception of Sesquiterpene lactones. Plant Cell Physiol. 64:996–1007. doi:10.1093/pcp/pcad026
- Teoh, K. H., Polichuk, D. R., Reed, D. W., and Covello, P. S. 2009. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. This paper is one of a selection of papers published in a Special Issue from the National Research Council of Canada – Plant Biotechnology Institute. Botany 87:635–642. doi:10.1139/B09-032
- Teoh, K. H., Polichuk, D. R., Reed, D. W., Nowak, G., and Covello, P. S. 2006. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580:1411–1416. doi:10.1016/j.febslet.2006.01.065
- Toda, Y., Okada, K., Ueda, J., and Miyamoto, K. 2019. Dehydrocostus lactone, a naturally occurring polar auxin transport inhibitor, inhibits epicotyl growth by interacting with auxin in etiolated Pisum sativum seedlings. Acta Agrobot. 72:doi:10.5586/aa.1779
- Toh, S., Holbrook-Smith, D., Stogios, P. J., Onopriyenko, O., Lumba, S., Tsuchiya, Y., Savchenko, A., and McCourt, P. 2015. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350:203–207. doi:10.1126/science.aac9476
- Tostes, J. B. F., Carvalho, A. L. D., Ribeiro da Silva, A. J., Mourão, P. J. P., Rossi, Á. D., Tanuri, A., and Siani, A. C. 2021. Phorbol esters from the latex of Euphorbia umbellata: bioguided isolation of highly potent HIV-1 latency interrupters in virus reservoir cells. J. Nat. Prod. 84:1666–1670. doi:10.1021/acs.jnatprod.0c01092
- Tsuchiya, Y., Yoshimura, M., Sato, Y., Kuwata, K., Toh, S., Holbrook-Smith, D., Zhang, H., McCourt, P., Itami, K., Kinoshita, T., and Hagihara, S. 2015. PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349:864–868. doi:10.1126/science.aab3831
- Tu, Y. 2016. Artemisinin - a gift from traditional Chinese medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55:10210–10226. doi:10.1002/anie.201601967
- Vahabi, K., Balcke, G. U., Hakkert, J. C., van der Meer, I., Athmer, B., and Tissier, A. F. 2024. Metabolome and transcriptome profiling of root chicory provide insights into laticifer development and specialized metabolism. bioRxiv 2024.2001.2002.573856.
- Varshney, K., and Gutjahr, C. 2023. KAI2 can do: Karrikin receptor function in plant development and response to abiotic and biotic factors. Plant Cell Physiol. 64:984–995. doi:10.1093/pcp/pcad077
- Vertrees, G. L., and Mahlberg, P. G. 1978. Structure and ontogeny of laticifers in Cichorium intybus (Compositae). Am. J. Botany 65:764–771. doi:10.1002/j.1537-2197.1978.tb06135.x
- Waegneer, E., Rombauts, S., Baert, J., Dauchot, N., De Keyser, A., Eeckhaut, T., Haegeman, A., Liu, C., Maudoux, O., Notté, C., Staelens, A., Van der Veken, J., Van Laere, K., and Ruttink, T. 2023. Industrial chicory genome gives insights into the molecular timetable of anther development and male sterility. Front. Plant Sci. 14:1181529. doi:10.3389/fpls.2023.1181529
- Wallaart, T. E., Bouwmeester, H. J., Hille, J., Poppinga, L., and Maijers, N. C. 2001a. Amorpha-4,11-diene synthase: cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin. Planta 212:460–465. doi:10.1007/s004250000428
- Weid, M., Ziegler, J., and Kutchan, T. M. 2004. The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proc. Natl. Acad. Sci. U S A. 101:13957–13962. doi:10.1073/pnas.0405704101
- Werker, E. 2000. Trichome diversity and development. Adv Botan Res. 31:1–35. doi:10.1016/S0065-2296(00)31005-9
- Wu, W., Huang, H., Su, J., Yun, X., Zhang, Y., Wei, S., Huang, Z., Zhang, C., and Bai, Q. 2022. Dynamics of germination stimulants dehydrocostus lactone and costunolide in the root exudates and extracts of sunflower. Plant Signal. Behav. 17:2025669–2025669. doi:10.1080/15592324.2022.2025669
- Wulfkuehler, S., Gras, C., and Carle, R. 2014. Influence of light exposure during storage on the content of sesquiterpene lactones and photosynthetic pigments in witloof chicory (Cichorium intybus L. var. foliosum Hegi). LWT Food Sci. Technol. 58:417–426. doi:10.1016/j.lwt.2014.04.017
- Xin, H., Ji, F., Wu, J., Zhang, S., Yi, C., Zhao, S., Cong, R., Zhao, L., Zhang, H., and Zhang, Z. 2023. Chromosome-scale genome assembly of marigold (Tagetes erecta L.): an ornamental plant and feedstock for industrial lutein production. Hortic. Plant J. 9:1119–1130. doi:10.1016/j.hpj.2023.04.001
- Yokotani-Tomita, K., Kato, J., Kosemura, S., Yamamura, S., Kushima, M., Kakuta, H., and Hasegawa, K. 1997. Light-induced auxin-inhibiting substance from sunflower seedlings. Phytochemistry 46:503–506. doi:10.1016/S0031-9422(97)00307-5
- Yokotani-Tomita, K., Kato, J., Yamada, K., Kosemura, S., Yamamura, S., Bruinsma, J., and Hasegawa, K. 1999. 8-epixanthatin, a light-induced growth inhibitor, mediates the phototropic curvature in sunflower (Helianthus annuus) hypocotyls. Physiol. Plant. 106:326–330. doi:10.1034/j.1399-3054.1999.106310.x
- Zabel, S., Brandt, W., Porzel, A., Athmer, B., Bennewitz, S., Schäfer, P., Kortbeek, R., Bleeker, P., and Tissier, A. 2021. A single cytochrome P450 oxidase from Solanum habrochaites sequentially oxidizes 7-epi-zingiberene to derivatives toxic to whiteflies and various microorganisms. Plant J. 105:1309–1325. doi:10.1111/tpj.15113
- Zakariya, A. M., Saiman, M. Z., Simonsen, H. T., and Ikram, N. K. K. 2023. Current state, strategies, and perspectives in enhancing artemisinin production. Phytochem. Rev. doi:10.1007/s11101-023-09897-6
- Zhang, Y., Teoh, K. H., Reed, D. W., Maes, L., Goossens, A., Olson, D. J. H., Ross, A. R. S., and Covello, P. S. 2008. The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of Artemisinin in Artemisia annua. J. Biol. Chem. 283:21501–21508. doi:10.1074/jbc.M803090200
- Zhang, L., F. Jing, F. Li, M. Li, Y. Wang, G. Wang, X. Sun and K. Tang (2009). Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol. Appl. Biochem. 52(3):199–207.
- Zhang, B., Wang, Z., Han, X., Liu, X., Wang, Q., Zhang, J., Zhao, H., Tang, J., Luo, K., Zhai, Z., Zhou, J., Liu, P., He, W., Luo, H., Yu, S., Gao, Q., Zhang, L., and Li, D. 2022. The chromosome-scale assembly of endive (Cichorium endivia) genome provides insights into the sesquiterpenoid biosynthesis. Genomics 114:110400–110400. doi:10.1016/j.ygeno.2022.110400