Abstract
Citrus melanose, caused by Diaporthe citri, is one of the most economically significant fungal diseases in China. Given that the application of fungicides is the main technique used for the control of citrus melanose, many studies have focused on enhancing the effectiveness and efficiency of the use of fungicides for the control of this economically significant disease. Here, we developed a TaqMan quantitative polymerase chain reaction (qPCR) protocol for monitoring citrus melanose via detection of suspected infected leaves and airborne conidia of D. citri. A primer pair and TaqMan probe targeting D. citri were designed to amplify a region of the fungal β-tubulin gene. The limits of detection and quantification of our TaqMan qPCR method were 159.57 and 683 copies/μL, respectively. The method detected the DNA equivalent to 47 conidia when fungal conidia were cultured in vitro. To evaluate the feasibility of our TaqMan qPCR method, symptomatic and asymptomatic leaf samples, airborne conidia samples were collected from a citrus orchard and tea garden, respectively. D. citri was detected in leaves with citrus melanose symptoms in an orchard; airborne fungal conidia of D. citri were also detected. These findings suggested that this novel TaqMan qPCR method could be used to detect D. citri both in vivo and in vitro. Our methodology could be applied as an alternative method for monitoring the changes in the incidence of D. citri in orchards.
Introduction
Citrus melanose is a fungal disease caused by Diaporthe citri F.A. Wolf [Citation1] that results in the formation of black and brown spots on the surface of citrus fruits, leaves and branches. Citrus fruit with a serious infection can have tears or greasy scabs. This has major deleterious effects on the quality of citrus fruit and can induce significant economic losses. Citrus melanose has been documented in citrus orchards worldwide [Citation2], especially in citrus-growing areas of the United States [Citation3], Pakistan [Citation4], Korea [Citation5,Citation6], Japan [Citation7], and Zhejiang Province [Citation8] and Jiangxi Province [Citation9] in China. The ascospores and conidia of D. citri, which are produced in deadwood and decaying branches, are causative agents citrus melanose. The pycnidium continuously releases conidia in deadwood [Citation10], which keeps the deadwood highly infectious for a long time. Thus, fungicides need to be applied frequently over long periods to effectively control the disease [Citation3,Citation11]. Copper hydroxide [Citation3] and mancozeb [Citation8,Citation12–14] are effective fungicides for the control of citrus melanose. The timing of the application of these fungicides is mainly based on the phenology of citrus plants [Citation3,Citation11] and rainfall [Citation13,Citation15]; that is, 250–300 mm of accumulated rainfall is used as a reference for the next spray. Given the variations in phenology and rainfall patterns, these factors cannot be used to reliably optimize the timing of fungicide applications. In some cases, fungicide application programs designed based on these factors can be far from optimal. There is thus a pressing need to identify more reliable factors that could be used for the development of improved fungicide application programs.
D. citri monitoring can provide valuable information on changes in the abundance of this pathogen in orchards, and this has major implications for the design of fungicide application regimes. Approaches that permit the rapid and accurate detection of D. citri are needed to effectively monitor this pathogen and clarify temporal patterns in the incidence of citrus melanose. Such approaches could provide useful information for the development of integrated prevention and control measures. Quantitative PCR (qPCR) is one of the most valuable tools in molecular biology research. It can be used to detect small amounts of DNA, and the limit of detection (LOD) and limit of quantification (LOQ) can be analysed using a discrete threshold approach and a curve-fitting modelling approach [Citation16].
Quantitative PCR (qPCR) has been used in diverse applications in an increasing number of recent agricultural studies [Citation17,Citation18]. The conidia of two important fungal pathogens, Leptosphaeria maculans and Pyrenopeziza brassicae, have been reported to be accurately detected when the DNA equivalent was 10 conidia in a PCR mixture [Citation19]. Other studies have shown that qPCR can be a valuable tool for the quantification of plant pathogens in the field, such as the causative agents of botrytis leaf blight of onion (Botrytis squamosa) [Citation17], blast fungus of rice (Pyricularia oryzae Cav.) [Citation20] and powdery mildew of peach (Podosphaera pannosa Wallr.) [Citation18]. Overall, these studies have suggested that qPCR can be a highly useful tool for monitoring of plant pathogens in the field.
Phenotypic characterizations of symptomatic leaves and fruits can be used for the detection of citrus melanose, but molecular techniques have also been developed. Conventional PCR and SYBR Green qPCR technologies have recently been used to verify the identity of D. citri mycelia and D. citri-infected citrus plants. The SYBR Green qPCR method is more sensitive than conventional PCR for the detection of D. citri [Citation21]. This indicates that qPCR might be effective for the rapid identification of citrus melanose in citrus orchards. Here, we developed a TaqMan qPCR method for detecting the causative agent of citrus melanose via the quantitative monitoring of airborne conidia. We first designed TaqMan qPCR primers and probes targeting regions of the beta-tubulin (TUB) gene. Next, conventional PCR was conducted in D. citri, Colletotrichum gloeosporioides, Diaporthe lithocarpus, Diplodia natalensis Evans, Guignardia citricarpa, Diaporthe nobilis, and asymptomatic citrus leaves to confirm the accuracy of the primers for the amplification of partial TUB sequences. The specific primers were used in future experiments to optimize the annealing temperature, prepare the standard curve and determine the sensitivity of the qPCR method. Finally, citrus leaves with symptoms of citrus melanose and airborne conidia were collected from orchards and used to verify the utility of the TaqMan qPCR method for the detection of D. citri. This method could be used to rapidly and accurately identify plants with citrus melanose symptoms, as well as to monitor the incidence of citrus melanose via quantification of airborne conidia in orchards.
Materials and methods
Fungal strains and mycelia DNA extraction
Nine fungal strains were used for TaqMan qPCR detection (), including three strains of D. citri(PP355401(the lhoh strain), PP355402, PP355405), two strains of Colletotrichum gloeosporioides(PP373722, PP373723), and one strain each of D. lithocarpus(PP355418), D. nobilis(PP355419), Diplodia natalensis (morphologically) and Guignardia citricarpa(PP355420). These strains are prevalent in Zhejiang Province, and they have been isolated from the organs of citrus plants in Taizhou and Quzhou, Zhejiang Province, China.
Table 1. Information on selected fungal strains in this study.
Fungal strains are collected and stored in Zhejiang Citrus Research Institute.
All fungal strains were cultivated on plates containing 15 mL of potato dextrose agar (PDA): 200 g/L potato extract, 20 g/L dextrose, 15 g/L agar, 1 L dH2O, pH = 7.0. After the plates were inoculated, they were maintained at 25 °C for seven days. Next, sterile loops were used to collect fungal mycelia, and the mycelia were ground in liquid nitrogen. The cetyltrimethylammonium bromide method was used to extract fungal DNA [Citation29], and an ultraviolet spectrophotometer and 1.5% agarose gel electrophoresis were used to determine the sample concentration and quality, respectively. The extracted DNA was stored at −20 °C for further analyses.
Primer and TaqMan probe design and verification
The nucleotide sequences of the β-tubulin gene (TUB: KJ490399, KC843187, KC357456, KJ490395, KJ490400, KJ490401, MH063914, MW208534 and MW836783) in D. citri were obtained from the GenBank database of the National Center for Biotechnology Information (https://www.ncbi.nlm. nih.gov/genbank/). The nucleotide sequences were aligned in MEGA 7.0 software, and the conserved regions were used in subsequent experiments [Citation30]. Primer Express v3.0 was used to design primer pairs (DcitTUB-F/DcitTUB-R) and TaqMan probes (DcitTUB-P) for TUB [Citation31] (). The Primer-BLAST tool in GenBank was used to verify the specificity of the primer pairs; all primers were specifically matched to the D. citri genome sequence. Conventional PCR was used to verify the efficiency of the primer pairs. The reaction system (25 μL) comprised 2.5 μL of 10 × Tap buffer, 2 μL of dNTP, 1.5 μL of each primer (10 μmol/L), 0.4 μL of DNA polymerase (2.5 U), 1 μL of fungal DNA, and 16.1 μL of double-distilled water (ddH2O). The thermal cycling conditions were as follows: 94 °C for 3 min; 35 cycles of 94 °C for 1 min, 59 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min. Conventional PCR was performed using an ETC821 system (Eastwin Scientific Equipments Inc.). The reaction was then cooled to 4 °C. PCR products were visualized in a 1.5% agarose gel.
Table 2. Primers and TaqMan probes designed in this study.
Optimization and verification of TaqMan qPCR
TaqMan qPCR was conducted using the Premix ExTaqTM PCR kit (Probe qPCR) (TaKaRa Dalian Co., Ltd.) in reactions containing the following: 10 μL of 2 × Premix Ex buffer, 0.5 μL of each primer (10 μmol/L), 0.5 μL of probe (10 μmol/L), 2 μL of fungal DNA, and 6.5 μL of ddH2O. The thermal cycling conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 52∼62 °C for 30 s. The optimal annealing temperature was determined using a temperature range of 52∼62 °C. TaqMan qPCR was performed using a CFX96 Real-Time PCR system (Bio-rad).
The LOD and LOQ for the TaqMan qPCR of D. citri were determined using the curve-fitting modelling method as described by Klymus et al. [Citation16]. Conventional PCR amplification was performed using the primer pair Bt2a/Bt2b, which are specific for the β-tubulin gene [Citation32,Citation33]. PCR products (541 bp) were recovered using a MiniBEST Agarose Gel DNA Extraction Kit (Code No. 9762, TaKaRa, Dalian Co., Ltd.). The recovered product was ligated to a pMD-19T Vector (2692 bp, TaKaRa, Dalian Co., Ltd.). The reaction system comprised 1 μL T-Vector pMD19, 1 μL PCR product, 3 μL ddH2O, 5 μL Solution I from the DNA Ligation Kit. Then the reactions were incubated for 30 min at 16 °C after mixing. The recombinant plasmid was transformed into (Escherichia coli) Trans1-T1 Phage Resistant Chenmically competent cells (Code No. CD501, TransGen Biotech Co., Ltd), and coated in LB medium containing ampicillin. After 24 h, colonies grew on the plate; single colonies were selected and extracted plasmids were subjected to PCR detection. For the purpose of conventional PCR, plasmids were extracted using the E.Z.N.ATM Plasmid DNA Mini Kit (Omega Bio-tek, Inc.). Sequencing of the recombinant plasmid in both directions was performed by Sangong Bioengineering (Shanghai Co., Ltd.).
The plasmid standard DNA concentration was diluted 10-fold to obtain 6 gradient concentrations; then these series of DNA templates were used for TaqMan qPCR analyses, and three replicate TaqMan qPCR reactions were performed for each concentration. The reaction system (20 μL) comprised 10 μL of 2 × Premix Ex buffer, 0.5 μL of each primer (10 μmol/L), 0.5 μL of probe (10 μmol/L), 2 μL of plasmid standard DNA, and 6.5 μL of ddH2O. After amplification, an R script (https://github.com/cmerkes/qPCR_LOD_Calc) was used to plot a curve of the standard concentration against Cq values and determine the LOD and LOQ.
Optimization of DNA extraction from fungal conidia
D. citri (the lhoh strain) was cultured on PDA plates until conidia were produced; the conidia were collected and diluted with TE buffer (10 mM Tris-HCl, 1 mmol/L EDTA, pH = 8.0). The concentration of conidia suspension was first adjusted to 5.4 × 107 per mL by a haemocytometer (XB-K-25, Shanghai Qiujing Biochemical Reagent Instrument Co., Ltd), and this was followed by successive dilutions to the following concentrations: 1.1 × 107, 2.1 × 106, 4.3 × 105, 8.6 × 104, 1.7 × 104, 3.4 × 103, 6.9 × 102 and 3.4 × 10. For each concentration, 100 μL of conidia suspension was transferred to three new 1.5 mL centrifuge tubes, which were placed into an automatic grinder (Jingxing Shanghai, Co., Ltd.) and ground five times at 60 Hz for 80 s. Four DNA extraction kits were used to determine the optimal kit for the extraction of DNA from conidia (). The extracted DNA was dissolved with 20 μL of TE buffer; 2 μL DNA was used for TaqMan qPCR. The four DNA extraction methods were hereafter referred to as Dingguo, Tiangen, Qiagen and Sangon.
Table 3. DNA extraction kits used for the extraction of DNA from conidia.
Application of the TaqMan qPCR method for the monitoring of orchards
To determine the efficiency of our TaqMan qPCR method for the monitoring of citrus melanose in the field, citrus leaves with melanose-like symptoms in the early stage and airborne conidia in the orchard were collected for DNA extraction and TaqMan qPCR quantification. A total of six leaf samples were collected from an orchard in Toutuo Town, Taizhou City, Zhejiang Province on 15 August 2023. Airborne conidia in an orchard (121°10’17.317″E, 28°38’49.632″N) and tea garden (121°10’2.834″E, 28°39’15.278″N) were collected using a TPBZ3 conidia trap (Zhejiang Tuo Pu Yun Nong Co., Ltd.) for 2 h a day from 15 to 16 August 2023. The samples were ground as described above, and the MagicMag Micro Genomic DNA Extraction Kit (Sangon) was used to extract microbial DNA. The extracted DNA was dissolved with 20 μL TE buffer, and 2 μL DNA was used for TaqMan qPCR. The mycelia and conidia of D. citri (the lhoh strain, 2.1 × 106 per mL) were used as the positive control; the tea garden sample and ddH2O were used as the negative control.
Results
Efficacy of the TaqMan qPCR method for the detection of fungal mycelia DNA
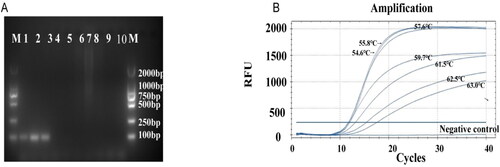
The primer pair DcitTUB-F/DcitTUB-R was used to amplify target DNA fragments from the genome of D. citri via conventional PCR; these primers did not successfully amplify target DNA fragments from the genomes of C. gloeosporioides, D. lithocarpus, D. nobilis, D. natalensis, G. citricarpa and asymptomatic leaves ((A)). The optimal annealing temperature was 57.6 °C for the primer pair DcitTUP-F/DciTUB-R ((B)). This was concluded based on the fact that the increase in the amplification of the target sequences occurred earlier at 57.6 °C compared with other temperatures. Furthermore, the log phase of the amplification curve was short at this temperature, and the plateau phase of the amplification curve was stable at 57.6 °C. Hence, an annealing temperature of 57.6 °C for DcitTUB-F/DcitTUB-R was used in subsequent TaqMan qPCR experiments.
Figure 1. Screening and optimization of D. citri primers. (A) Specific amplification via the primers DcitTUB-F/DcitTUB-R for D. citri; agarose gel electrophoresis parameters: 25 min, 110 V, 1.5% agarose gel. M: DL2000 DNA Marker (TaKaRa, Dalian Co., Ltd.); Lanes 1–3: D. citri; Lane 4: Colletotrichum gloeosporioides; Lane 5: C. gloeosporioides; Lane 6: D. lithocarpus; Lane 7: Diplodia natalensis Evans; Lane 8: Guignardia citricarpa; Lane 9: D. nobilis; Lane 10: Asymptomatic citrus leaves. (B) Curves for determination of the optimal annealing temperature for the primer pair DcitTUB-F/DcitTUB-R.
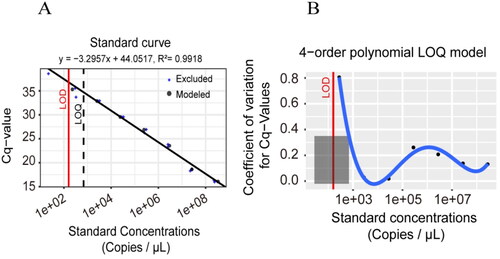
Next, the LOD and LOQ of our TaqMan qPCR method were determined. The DNA length of plasmid standard was 541 bp. Its concentration was determined by microspectrophotometer as 97.82 ng/μL, with OD260/OD280 = 1.87. Using the following formula, the copy number concentration was calculated to be 2.76 × 1010 copies/μL, copies/μL= (6.02 × 1023) × (ng/μL × 10−9)/(DNA length +2962)/660. When the plasmid standard concentration of D. citri was 2.76 × 108, 2.76 × 107, 2.76 × 106, 2.76 × 105, 2.76 × 104, 2.76 × 103 and 2.76 × 102 copies/μL, the Cq value was 16.12 ± 0.16, 18.52 ± 0.20, 23.53 ± 0.29, 26.69 ± 0.37, 29.53 ± 0.03, 32.88 ± 0.04 and 34.84 ± 1.02, respectively (y = 44.0517 − 3.2957x, R2=0.99, p < .05, (A)). The LOD and LOQ were determined to be 159.57 and 683 copies/μL, respectively, according to the curve-fitting method (). Thus, our TaqMan qPCR method was effective for detecting and quantifying low concentrations of target DNA.
Optimal for the extraction of DNA from fungal conidia
The Sangon method, which involves the use of Proteinase K lysis, was optimal for the extraction of DNA from fungal conidia (). When the concentration of conidia was greater than 8.6 × 104 per mL, qPCR succeed to detect the fungal DNA that had been extracted via all four methods. However, when the conidia concentration was less than 8.6 × 104 per mL, but greater and equal (≥) to 6.9 × 102, only the DNA that had been extracted using the Sangon method was detected by our qPCR method. Thus, the Sangon method was considered the most appropriate for further extractions of fungal conidia DNA.
Table 4. Comparison of different methods for the extraction of DNA from fungal conidia.
TaqMan qPCR method for the detection of fungal conidia DNA
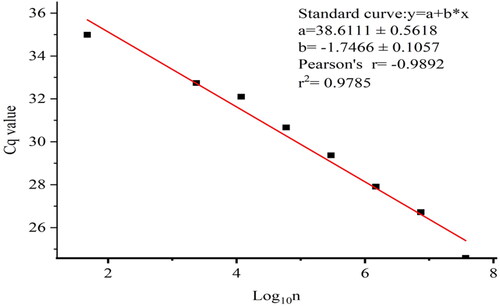
When the total number of conidia was 3.69 × 106, 7.38 × 105, 1.48 × 105, 2.95 × 104, 5.9 × 103, 1.18 × 103, 2.36 × 102 and 4.72 × 101, respectively, and the Cq value was 24.59, 26.72, 27.91, 29.37, 30.67, 32.1, 32.74 and 34.99 (y = 38.6111 − 1.7466x, R2=0.97, p < .05, ), the method was able to detected the DNA equivalent of 47 conidia. This indicates that our TaqMan qPCR method could be used to determine the number of fungal conidia.
Application of the TaqMan qPCR method for the detection of D. citri in orchard samples
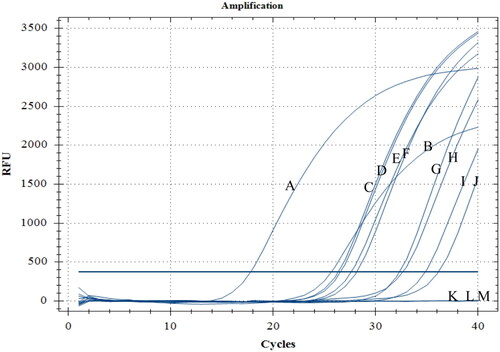
Our TaqMan qPCR method was used to detect D. citri in putatively infected leaves and quantify airborne conidia derived from D. citri in orchards. Six leaf samples and two conidia samples were used to evaluate the practical utility of our TaqMan qPCR method for the detection of D. citri (). D. citri was detected in all six leaf samples, and their Cq values ranged from 26.34 to 32.31. D. citri was also detected in the two airborne conidia samples, and their Cq values were 34.85 (142.35 conidia) and 36.13 (26.33 conidia).
Figure 4. TaqMan qPCR detection of leaf samples with symptoms of citrus melanose and airborne conidia from an orchard. A and B: positive control; C–H: leaf samples; I and J: airborne conidia captured using a TPBZ3 conidia trap in a citrus orchard; K: ddH2O; L and M: airborne conidia captured using a TPBZ3 conidia trap in a tea garden.
Discussion
We developed a new TaqMan qPCR method for the detection of the causative agent of citrus melanose: D. citri. We tested this method on both the fungal mycelia and conidia of D. citri and found that this method could be used to detect trace amounts of fungal DNA with high specificity and sensitivity. We also tested this method on field samples and detected D. citri in leaves showing symptoms of citrus melanose and airborne conidia from orchards. The LOD and LOQ of our TaqMan qPCR method were 159.57 and 683 copies/μL, respectively (), and the method was able to detect the DNA equivalent of 47 conidia (). qPCR method is more sensitive for the detection of D. citri than conventional PCR [Citation21]. The detection sensitivity of the designed D. citri specific primers in conventional PCR and qRT-PCR amplification was 0.78 ng/μL and 0.35 ng/μL, respectively. TaqMan qPCR has previously been shown to detect animal and plant pathogens more rapidly and with higher specificity, sensitivity and accuracy compared with other methods.
This qPCR method for DNA extracted from fungal conidia has various uses in the field of disease epidemiology, such as disease monitoring and forecasting [Citation34,Citation35]. The concentration of conidia obtained in the field has been used to forecast disease outbreaks and make fungicide application decisions, such as ergot disease in grass [Citation36]; powdery mildew of grape [Citation17]; Sclerotinia stem rot of rape [Citation37]; wheat rust, wheat smut, and wheat blight [Citation38]; powdery mildew of peach [Citation18]; and powdery mildew of wheat [Citation39]. The main method used for forecasting outbreaks of citrus melanose is based on phenotypic observations of symptomatic leaves and fruits. However, citrus melanose outbreaks cannot be forecasted in a timely manner using this method, which makes it of little accurately for the design of fungicide application regimes [Citation11,Citation40,Citation41]. Conidia have traditionally been quantified using microscopic counts, which are time-consuming. Our method provides an effective alternative to traditional methods; it permits citrus samples with symptoms of citrus melanose to be identified and can be used to monitor and forecast citrus melanose outbreaks via the quantification of D. citri conidia.
The concentration of aerial spores can be used to identify optimal thresholds for the application of fungicides. In previous studies, 8.548 sporangia per cubic metre [Citation42] and 10 spores [Citation43] were used as thresholds for the application of fungicides for the control of Bremia lactucae and Erysiphe necator, respectively. These thresholds can be used to adjust fungicide application intervals for the control of lettuce downy mildew and grape powdery mildew and significantly reduce the use of fungicides. The TaqMan qPCR procedure established in our study can detect DNA equivalent to 47 D. citri conidia, indicating that this technique would be effective for monitoring the airborne conidia of D. citri. This method could be used to clarify the disease epidemiology of citrus melanose and optimize the application interval of fungicides based on the concentration of airborne conidia.
Citrus melanose is most prevalent from April to October. The application of fungicides every half month during this period has been recommended in a previous study despite the lack of any information on the disease dynamics of citrus melanose [Citation11]. Epidemiological studies of the airborne conidia of D. citri will aid the control of this disease. Our method can be used to accurately quantify variation in the abundance of pathogens in orchards and optimize the timing of fungicide application.
Conclusions
In this study, a specific primer pair and TaqMan probe for D. citri were designed based on the β-tubulin gene, and a rapid molecular detection system using TaqMan qPCR was established. The symptomatic, asymptomatic leaf samples and airborne conidia samples were detected and verified. The TaqMan qPCR protocol exhibited high specificity, sensitivity and accuracy, which can be used for molecular diagnosis and dynamic detection of D. citri.
Author’s contributions
Conceived, designed the experiments, writing-original draft preparation, writing-review and editing, Z.X.P., F.H., Z.D.H.; Investigation, performed the experiments, sampling, Z.X.P., L.M.L., D.C.D., L.Z., X.R.H, Z.D.H.; Analysed the data, Z.X.P., Z.D.H. and L.M.L. Funding acquisition, Z.X.P., Z.D.H.; All authors have read and approved the manuscript.
Acknowledgements
The authors would like to thank Sangong Bioengineering (Shanghai Co., Ltd.) for provision of sequencing of the recombinant plasmid.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data that support the findings reported in this study are available from the corresponding author(Z.D.H.)upon reasonable request.
Additional information
Funding
References
- Wolf FA, Bach WJ. The isolation of the fungus that causes citrus melanose and the pathological anatomy of the host. J Agric Res. 1928;37:1–10.
- Chaisiri C, Liu X, Lin Y, et al. Diaporthe citri: a fungal pathogen causing melanose disease. Plants. 2022;11(12):1600. doi: 10.3390/plants11121600.
- Timmer LW, Zitko SE, Albrigo LG. Split applications of copper fungicides improve control of melanose on grapefruit in Florida. Plant Dis. 1998;82(9):983–986. doi: 10.1094/PDIS.1998.82.9.983.
- Rehman FU, Kalsoom M, Sultan A. Citrus melanose and quality degradation of fruit by this disease: a review. JBGSR. 2020;39(5):1–4. doi: 10.46718/JBGSR.2020.03.000081.
- Hyun JW, Yi PH, Hwang RY, et al. Aspect of incidence of the major citrus diseases recently. Res Plant Dis. 2013;19:102–107. doi: 10.5423/RPD.2013.19.2.102.
- Shin YH, Ko EJ, Kim SJ, et al. Suppression of melanose caused by Diaporthe citri on citrus leaves pretreated with bio-sulfur. Plant Pathol J. 2019;35(5):417–424. doi: 10.5423/PPJ.OA.03.2019.0067.
- Yutaka A, Yasuo H, Tomomasa M. Studies on citrus melanose and citrus stem-end rot by Diaporthe citri Wolf. Jpn J Phytopathol. 1986;52:39–46.
- Jiang LY, Xu FS, Huang ZD, et al. Occurrence and control of citrus melanose caused by diaporthe citri. Acta Agric Zhejiangensis. 2012;24(4):647–653. doi: 10.3969/j.issn.1004-1524.2012.04.022.
- Zeng BL, Zeng ZF, Zhu XY, et al. Occurrence regularity and comprehensive control measures of nanfeng orange fruit black spot disease. Zhejiang Ganju. 2010;27(20):24–26. doi: 10.3969/j.issn.1009-0584.2010.02.009.
- Mondal SN, Agostini JP, Zhang L, et al. Factors affecting pycnidium production of Diaporthe citri on detached citrus twigs. Plant Dis. 2004;88(4):379–382. doi: 10.1094/PDIS.2004.88.4.379.
- Huang XB, Kh L, Huang ZD. Study on the suitable time and medication method of controlling citrus black spot disease. Zhejiang Ganju. 2010;27(2):26–29. doi: 10.3969/j.issn.1009-0584.2010.02.010.
- Chen GQ, Jiang LY, Fs X, et al. In vitro and in vivo screening of fungicides for controlling citrus melanose caused by Diaporthe citri. J Zhejiang Univ Agric Life Sci. 2010;36(4):440–444. doi: 10.4028/www.scientific.net/AMM.37-38.1549.
- Noguchi M. Appropriate period control of citrus melanose using simple rain gauge. Fruit Jpn. 2013;68(8):53–55 [Japanese]. Available from: https://ndlsearch.ndl.go.jp/books/R000000004-I024777105
- Yi PH, Hyun JW, Hwang RY, et al. Improvement of control efficacy of mancozeb wettable powder against citrus melanose by mixing with paraffin oil. Res Plant Dis. 2014;20(3):196–200. doi: 10.5423/RPD.2014.20.3.196.
- Miyoshi T, Kawahata Y, Shimizu S. The effect of mancozeb wp (wettable powder) on mancozeb adhesion and melanose control after application on wet citrus trees. Jpn J Phytopathol. 2007;73(3):149–154. doi: 10.3186/jjphytopath.73.149.
- Klymus KE, Merkes CM, Allison MJ, et al. Reporting the limits of detection and quantification for environmental DNA assays. Environ DNA. 2020;2(3):271–282. doi: 10.1002/edn3.29.
- Carisse O, Tremblay DM, Lévesque CA, et al. Development of a TaqMan real-time PCR assay for quantification of airborne conidia of Botrytis squamosa and management of botrytis leaf blight of onion. Phytopathology. 2009;99(11):1273–1280. doi: 10.1094/PHYTO-99-11-1273.
- Marimon N, Eduardo I, León M, et al. A qPCR-based method for the detection and quantification of the peach powdery mildew (Podosphaera pannosa) in epidemiological studies. Eur J Plant Pathol. 2020;158(4):1005–1016. doi: 10.1007/s10658-020-02136-0.
- Calderon C, Ward E, Freeman J, et al. Detection of airborne inoculum of Leptosphaeria maculans and Pyrenopeziza brassicae in oilseed rape crops by polymerase chain reaction (PCR) assays. Plant Pathol. 2002;51(3):303–310. doi: 10.1046/j.1365-3059.2002.00721.x.
- Xu LY, Zhao LW, Hu YF, et al. Development of a qPCR detection method for monitoring conidial density of rice blast fungus in the air. Acta Agric Zhejiangensiss. 2016;28(8):1368–1373. doi: 10.3969/j.issn.1004-1524.2016.08.14.
- Zeng YT, Xiong T, Li HY. Rapid molecular detection of Diaporthe citri, the pathogen of citrus melanose. Acta Agric Zhejiangensis. 2022;34(7):1457–1465. doi: 10.3969/j.issn.1004-1524.2022.07.13.
- Huang F, Hou X, Dewdney MM, et al. Diaporthe species occurring on citrus in China. Fungal Divers. 2013a;61(1):237–250. doi: 10.1007/s13225-013-0245-6.
- Huang F, Chen GQ, Hou X, et al. Colletotrichum species associated with cultivated citrus in China. Fungal Divers. 2013b;61(1):61–74. doi: 10.1007/s13225-013-0232-y.
- Luo M, Dong ZY, Bin SY, et al. First report of fruit rot disease on pomelo caused by Lasiodiplodia theobromae in China. Plant Dis. 2011;95(9):1190–1190. doi: 10.1094/PDIS-03-11-0214.
- Phillips AJL, Alves A, Abdollahzadeh J, et al. The Botryosphaeriaceae: genera and species known from culture. Stud Mycol. 2013;76(1):51–167. doi: 10.3114/sim0021.
- Wulandari NF, To-Anun C, Hyde KD, et al. Phyllosticta citriasiana sp. nov., the cause of citrus tan spot of citrus maxima in Asia. Fungal Divers. 2009;34:23–39.
- Li Y, Tan P, Zhao DG. Diaporthe nobilis, a new record on camellia sinensis in Guizhou province, China. Mycosphere. 2017;8(1):1–8. doi: 10.5943/mycosphere/8/1/1.
- Gao YH, Su YY, Sun W, et al. Diaporthe species occurring on lithocarpus glabra in China, with descriptions of five new species. Fungal Biol. 2015;119(5):295–309. doi: 10.1016/j.funbio.2014.06.006.
- Bartlett JMS, Stirling D. DNA extraction from fungi, yeast, and bacteria. Methods Mol Biol. 2003;226:53. doi: 10.1385/1-59259-384-4:53.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Rodríguez A, Rodríguez M, Córdoba JJ, et al. Design of primers and probes for quantitative real-time PCR methods. Methods Mol Biol. 2015;1275:31–56. doi: 10.1007/978-1-4939-2365-6_3.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.
- Udayanga D, Liu X, Crous PW, et al. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers. 2012;56(1):157–171. doi: 10.1007/s13225-012-0190-9.
- Van der Heyden H, Dutilleul P, Charron JB, et al. Monitoring airborne inoculum for improved plant disease management. A review. Agron Sustain Dev. 2021;41(3):1–23. doi: 10.1007/s13593-021-00694-z.
- Yan ZY, Fan JR, Liu W, et al. Models of disease index estimation of wheat powdery mildew based on the concentrations of Blumeria graminis f.sp. tritici conidia in the fields. Acta Phytopathol Sin. 2017;47(2):253–261.
- Jeremiah KS, Jeness C, Cheng Q, et al. Detection and quantification of airborne Claviceps purpurea sensu lato ascospores from hirst-type spore traps using real-time quantitative PCR. Plant Dis. 2018;102(12):2487–2493. doi: 10.1094/PDIS-02-18-0310-RE.
- Rogers SL, Atkins SD, West JS. Detection and quantification of airborne inoculum of Sclerotinia sclerotiorumusing quantitative PCR. Plant Pathol. 2009;58(2):324–331. doi: 10.1111/j.1365-3059.2008.01945.x.
- Araujo GT, Amundsen E, Frick M, et al. Detection and quantification of airborne spores from six important wheat fungal pathogens in Southern Alberta. Can J Plant Pathol. 2021;43(3):439–454. doi: 10.1080/07060661.2020.1817795.
- Cao X, Yao D, Zhou Y, et al. Detection and quantification of airborne inoculum of Blumeria graminisf.s p. tritici using quantitative PCR. Eur J Plant Pathol. 2016;146(1):225–229. doi: 10.1007/s10658-016-0908-8.
- Cai ZG, Zhuang ZY. Citrus melanose and its control. ZhongGuo Yuan Yi (Horticulture of Chinese). 1985;31(3):132–140.
- Liu X, Wang MS, Mei XF, et al. Sensitivity evaluation of Diaporthe citri populations to mancozeb and screening of alternative fungicides for citrus melanose control. J Plant Protect. 2018;45(2):373–381. doi: 10.13802/j.cnki.zwbhxb.2018.2016175.
- Dhar N, Mamo BE, Subbarao KV, et al. Measurements of aerial spore load by qPCR facilitates lettuce downy mildew risk advisement. Plant Dis. 2019;104(1):82–93. doi: 10.1094/PDIS-03-19-0441-RE.
- Thiessen LD, Neill TM, Mahaffee WF. Timing fungicide application intervals based on airborne Erysiphe necator concentrations. Plant Dis. 2017;101(7):1246–1252. doi: 10.1094/PDIS-12-16-1727-RE.