ABSTRACT
Introduction
Mitochondrial LonP1 is an ATP-powered protease that also functions as an ATP-dependent chaperone. LonP1 plays a pivotal role in regulating mitochondrial proteostasis, metabolism and cell stress responses. Cancer cells exploit the functions of LonP1 to combat oncogenic stressors such as hypoxia, proteotoxicity, and oxidative stress, and to reprogram energy metabolism enabling cancer cell proliferation, chemoresistance, and metastasis.
Areas covered
LonP1 has emerged as a potential target for anti-cancer therapeutics. We review how cytoprotective functions of LonP1 can be leveraged by cancer cells to support oncogenic growth, proliferation, and survival. We also offer insights into small molecule inhibitors that target LonP1 by two distinct mechanisms: competitive inhibition of its protease activity and allosteric inhibition of its ATPase activity, both of which are crucial for its protease and chaperone functions.
Expert opinion
We highlight advantages of identifying specific, high-affinity allosteric inhibitors blocking the ATPase activity of LonP1. The future discovery of such inhibitors has potential application either alone or in conjunction with other anticancer agents, presenting an innovative approach and target for cancer therapeutics.
1. Introduction
Oncogenic transformation of somatic cells is driven by a complex interplay of genetic and epigenetic alterations, which either activate oncogene-dependent pathways or deactivate tumor-suppressive mechanisms. Genotoxic stress often triggers replication stress, which is a critical initiator of tumorigenesis in its early stages. Furthermore, aneuploidy frequently arises early in tumorigenesis, disrupting the balance of proteins and promoting proteotoxicity caused by protein misfolding, misassembly and aggregation. In their quest to proliferate and survive, cancer cells must overcome other cellular stressors, which include hypoxia, oxidative and metabolic stress, amongst others. Cancer cells meet these challenges by adaptively reprogramming normal cytoprotective mechanisms, to sustain and increase their oncogenic potential.
Mitochondrial LonP1 has emerged as a potential target for anti-cancer therapeutics. It belongs to the superfamily of ATPases Associated with diverse cellular Activities (AAA+), which govern various cellular processes including DNA replication, gene expression, membrane fusion, and protein degradation [Citation1]. Within human mitochondria, there are four AAA+ proteases- LonP1 and ClpXP are soluble complexes in the matrix, while m-AAA and i-AAA are transmembrane complexes with active sites in the matrix and intermembrane space, respectively [Citation1]. LonP1 plays versatile roles in regulating cellular homeostasis and mediating adaptive responses to cell stress. Here, we review how the cytoprotective functions of LonP1 are likely exploited by cancer cells to promote their oncogenic growth, proliferation, and survival. We provide perspective on recently identified small molecule inhibitors of LonP1 that allosterically inhibit its ATPase activity necessary for protease and chaperone-like functions. We suggest that a novel and promising therapeutic approach lies in targeting the allosteric inhibition of ATP hydrolysis by LonP1 and other mitochondrial matrix AAA+ proteases. Administering these inhibitors, either as standalone treatments or in combination to potentiate existing anti-cancer chemotherapeutics, may be an innovative avenue for effectively combating cancer.
2. LonP1 in cellular homeostasis and cell stress responses
Human LonP1 is highly conserved throughout evolution from archaea, eubacteria to eukaryotes. In mice, the homozygous deletion of LonP1 causes early embryonic lethality [Citation2]. In eukaryotes, LonP1 is located in the mitochondrial matrix where it preserves cellular homeostasis and responds to cell stress [Citation3] ().
Figure 1. Functions of LonP1 in cellular homeostasis and cell stress responses that can be exploited by cancer cells (created using BioRender.com).
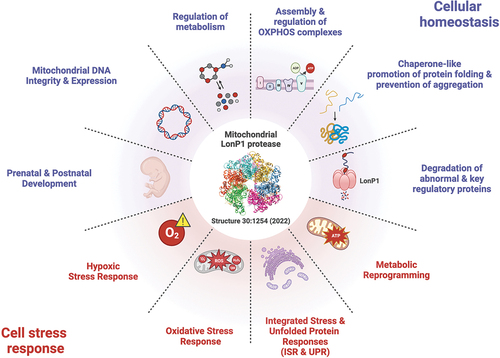
2.1. Multifaceted roles of LonP1 in the maintenance of mitochondrial homeostasis
LonP1 plays diverse roles in regulating mitochondrial proteostasis, metabolism and cell stress responses. As a protein quality control protease, it degrades misfolded, misassembled, and oxidatively damaged proteins [Citation3]. As a regulator of mitochondrial metabolism LonP1 also degrades key rate-limiting proteins involved, for example, in cholesterol metabolism, heme biosynthesis, and mitochondrial transcription. LonP1 degrades: 1) StAR, the steroidogenic acute regulatory protein, which mediates the rate-limiting transfer of cholesterol from the mitochondrial outer membrane to the inner membrane [Citation4]; 2) ALAS-1, delta-aminolevulinate synthase 1, which catalyzes the rate-limiting step in heme biosynthesis [Citation5]; and 3) TFAM, transcription factor A of mitochondria, which is required for the activation of mitochondrial transcription and the maintenance of mitochondrial DNA (mtDNA) [Citation6]. LonP1 also functions as an ATP-dependent chaperone, independent of its proteolytic activity, which can promote protein folding and assembly, and prevent aggregation. LonP1 is required for the proper folding of mitochondrial Hsp70 and its cognate co-chaperone DNAJA3/Tid1 [Citation7]. LonP1 helps maintain the solubility of newly synthesized polypeptides translocated into the mitochondrial matrix, thereby preventing aggregation and supporting proper folding through its ATPase (not protease) activity [Citation8]. The chaperone activity of LonP1 has been implicated in facilitating the assembly and regulation of oxidative phosphorylation (OXPHOS) complexes [Citation9].
As a regulator of cellular metabolism, LonP1 is critical for modulating pyruvate dehydrogenase (PDH) [Citation10,Citation11], the gatekeeper linking glycolysis with the TCA cycle. In isolated mouse heart mitochondria, LonP1 degrades PDH kinase 4 (PDK4) [Citation10], preventing the phosphorylation of the PDH E1α subunit, which inhibits PDH. In human fibroblasts, LonP1 also degrades phosphoE1α, providing another mechanism by which it regulates PDH activity [Citation11]. Together, these data suggest that LonP1 fine-tunes glucose and fatty acid oxidation by modulating the phosphorylation and activity of PDH.
Additionally, LonP1 has a DNA-binding property that is conserved from bacteria to humans [Citation12–14]. In human cells and worms, LonP1 binds mtDNA within the non-coding region [Citation13,Citation15] where replication and transcription are initiated. It plays a critical role in maintaining the integrity and expression of mtDNA, which encode essential subunits of OXPHOS complexes, as well as tRNAs and rRNAs vital for mitochondrial translation [Citation6,Citation13]. Notably, the embryonic demise of homozygous LonP1 null mice coincides with a substantial decline in mtDNA copy number [Citation2].
LonP1 is crucial during pre- and post-natal development (). Homozygous, compound heterozygous, and de novo mutations in the LONP1 gene cause a broad spectrum of rare developmental diseases, which can affect multiple organ systems as in CODAS syndrome, characterized by cerebral, ocular, auricular, dental, and skeletal anomalies [Citation16,Citation17]. Dysfunctional LonP1 in CODAS syndrome may explain the ocular and skeletal abnormalities, which are the key clinical hallmarks of this disease. LonP1 may be indispensable for protecting tissues during cartilage formation and bone ossification where ROS-dependent signaling is heightened, and in interior regions of the eye that are avascular and relatively hypoxic.
2.2. Roles of LonP1 in cell stress responses
LonP1 mitigates cell stress in several ways (). During hypoxia, low oxygen up-regulates LonP1, which facilitates the remodeling of the cytochrome c oxidase complex (COX) in the electron transport chain (ETC) [Citation18]. Hypoxia-inducible factor-1 alpha (HIF-1α), activates the transcription of LonP1, and increased levels of LonP1 degrade the COX subunit 4–1 [Citation18]. At the same time, HIF-1α activates the expression of an alternate subunit COX4–2, which is assembled into the COX complex. This subunit switching has been proposed to optimize the efficiency of the ETC to hypoxic conditions [Citation18].
In response to oxidative stress, LonP1 degrades oxidized proteins, and also employs its protease and chaperone activities to abrogate deleterious processes that threaten cell survival. A study in neuroblastoma and HeLa cell lines showed that LonP1 functions in concert with ClpXP to extinguish ROS in mitochondria by degrading the domain of Complex I of the ETC, which generates ROS [Citation19]. Another study using colon cancer cells and mouse embryonic fibroblasts showed that elevated ROS prompts p53 translocation from the cytosol into the mitochondrial matrix triggering necrosis [Citation20]. Similarly, experiments using oral squamous carcinoma and HEK293(T) cell lines showed that oxidative stress leads to p53 accumulation in the matrix, resulting in cytochrome c release and apoptosis [Citation21]. However, overexpression of wild-type LonP1 almost completely abrogated cytochrome c release and apoptosis, whereas a chaperone mutant of LonP1 (K529R) failed to block apoptosis elicited by oxidative stress [Citation21].
LonP1 participates in the integrated stress response (ISR) as well as the unfolded protein response in the endoplasmic reticulum (UPRER) and mitochondria (UPRmt) [Citation22,Citation23]. UPRER and UPRmt are characteristically induced by misfolded and misassembled proteins in these respective organelles, whereas the ISR is induced by amino acid deprivation, viral infection, heme deficiency, and other cellular stressors including ER stress. Although the knockout or knockdown of yeast and mammalian Lon homologs lead to mitochondrial dysmorphology and the accumulation of electron dense inclusions within mitochondria, which are often large and extensive, organisms and cells continue to proliferate often comparably to corresponding controls and do not die prematurely [Citation16,Citation24]. What remains unanswered is whether these inclusions are proteinaceous or elemental in nature, and whether they represent a cytoprotective response by sequestering toxic forms of proteins and elements (e.g., calcium, iron, phosphate).
A role for LonP1 in regulating metabolic flexibility is demonstrated by its selective degradation of PDK4 [Citation10], a component of the PDH complex bound to the E2 subunit [Citation10]. Metabolic flexibility is the ability of cells to generate ATP by preferentially using glucose or lipids (e.g. fatty acids and ketone bodies). For instance, in a cardiac muscle cell line cultured with glucose, LonP1 rapidly degrades PDK4 [Citation10]. However, when these cells are cultured with fatty acids, PDK4 degradation is suppressed leading to phosphorylation of PDH E1α and PDH inhibition [Citation10]. Additionally, in fibroblasts cultured in glucose, LonP1 degrades phosphoE1α thereby preventing PDH inhibition [Citation11]. Further work is required to understand the coordinated roles of LonP1 in regulating metabolic flexibility.
3. Exploitation of LonP1 in cancer: a survival strategy
Cancer cells face various oncogenic stressors like hypoxia, oxidative, proteotoxic, and metabolic stress, which align with processes influenced by LonP1 (, red shading). In leukemia cell lines, upregulated LonP1 degrades COX4–1 favoring COX4-2 assembly, which adapts OXPHOS efficiency to hypoxia [Citation18,Citation25]. The same study also showed increased levels of PDH kinase 1 (PDK1) during hypoxia [Citation25], raising the possibility that modulation of PDK1 degradation by LonP1 may be involved. Additionally, in prostate adenocarcinoma cells, LonP1 and ClpP (the proteolytic component of ClpXP) mitigate oxidative and proteotoxic stress as the knockdown of both AAA+ proteases increased production of mitochondrial ROS, accumulation of aggregated and misfolded proteins, and induced cell death [Citation26]. A key hallmark of cancer cells is the reprogramming of metabolism from OXPHOS to aerobic glycolysis (i.e. Warburg effect). One mechanism promoting this metabolic switch is via PDH inhibition. LonP1 may be involved in mediating this shift by increased degradation of PDH phosphatases (PDPs) and decreased degradation of PDKs and phosphoE1α; studies are required to test this possibility. Alternatively, a study in a prostate cancer cell line subjected to hypoxia, showed that LonP1 safeguards OXPHOS. LonP1 was phosphorylated by Akt (aka protein kinase B), leading to its enhanced protein quality control of OXPHOS complexes, and dampened ROS generation, resulting in increased tumor cell migration and invasion [Citation27].
4. LonP1 expression in cancer cells
4.1. Normal tissues- proteomics and transcriptomics
According to Human Protein Atlas (HPA) proteomics data, LonP1 is constitutively expressed throughout most tissues of the body. The HPA uses an immunohistochemistry approach to annotate protein expression in situ using 44 normal tissue types from 144 individuals [Citation28]. High protein expression levels are seen in the adrenal glands, gastrointestinal tract (appendix, colon, duodenum, rectum), gallbladder, and kidneys. The highest RNA expression is seen in adrenal gland and liver tissues based on the HPA and the Genotype-Tissue Expression (GTEx) RNA-Seq datasets [Citation28,Citation29].
4.2. Cancer cell lines and tissues - proteomics and transcriptomics
HPA cancer proteomics data (216 cancer patients, 20 different cancer types) show most malignant tissues displayed immunoreactivity to anti-LonP1 antibodies, with the highest expression in colon/rectum, prostate, head/neck, and liver [Citation28]. Transcriptomics data from The Cancer Genome Atlas (TCGA) (>20,000 primary cancer and matched normal samples across 33 cancer types) show LonP1 expression across all TCGA cancer types. In matched samples, increased LonP1 transcripts were seen in cervical squamous cell carcinoma, cholangiocarcinoma, colon adenocarcinoma, and esophageal carcinoma. Although higher levels of LonP1 are not prognostic, they are associated with lower survival in multiple cancers, including low-grade glioma and invasive breast carcinoma [Citation30].
4.3. Mutation rate
Alterations in the LONP1 gene are seen in 5.54% of cases profiled in the 2020 TCGA pan-cancer analysis of whole genomes, and only 2.64% were amplifications [Citation30].
4.4. LonP1 Isoforms
Recently in 2022, one study [Citation31] investigated the differential expression of three alternatively spliced LonP1 isoforms (ISO −1, −2, and −3) across cancers based on the TGCA Splicing Variants Database (TSVdb). To our knowledge, this is the only study that has addressed the expression of these LonP1 isoforms. ISO1 (959 aa) contains the complete amino-terminal mitochondrial targeting sequence (MTS); ISO2 (859 aa), has a truncated MTS; and ISO3 (763 aa), lacks an MTS. As compared to normal tissues, transcripts of ISO1 were significantly increased in 4 of the 9 cancer types analyzed - lung, prostate, breast, and bladder; while ISO2 were significantly elevated in 8 of 9 cancer types - prostate, breast, colon, rectum, cervical, head/neck, bladder, and renal. By contrast, transcripts of ISO3 were undetected in normal tissues except for head/neck and bladder and showed increased expression in 6 of 9 cancer types - breast, rectum, cervical, head/neck, bladder, and renal. To determine their subcellular localization, GFP fusion proteins were expressed in colorectal cancer cells; ISO1 was exclusively in mitochondria, ISO2 in mitochondria and cytoplasm, and ISO3 exclusively in the cytosol. Further work is needed to determine whether ISO2 and ISO3 have ATPase, protease, and chaperone-like activities. Future studies examining LonP1 must be cognizant of these isoforms.
5. Small molecule inhibitors of LonP1
Only a handful of small molecules have been shown to inhibit LonP1 but none of these are specific or of high affinity. Bortezomib and MG262 are dipeptide boronic acid inhibitors of the proteasome [Citation32], which also inhibit LonP1 at substantially lower efficacy [Citation33,Citation34]. When comparing these inhibitors in parallel, bortezomib inhibited the peptidase activity of purified human LonP1 with an IC50 of 17 nM, which is ~ 7 orders of magnitude lower activity than that of the 20S proteasome with an IC50 of 2.3 nM [Citation6] (). The cryoEM structure of bortezomib bound to human LonP1 shows that it binds at the proteolytic active site [Citation38]. Bortezomib is used clinically to treat multiple myeloma and mantle cell lymphoma [Citation32]. As the knockdown of LonP1 in mantle cell lymphoma cells leads to cell death [Citation24], it is unknown whether the inhibition of LonP1 by bortezomib might be of therapeutic benefit in treating hematologic malignancies. One study showed that in multiple myeloma cells, increased LonP1 levels resulted in decreased efficacy of proteasome inhibitors, conferring partial resistance [Citation39]. This suggests that the simultaneous inhibition of the proteasome and LonP1 by a single compound or by the combination of protease-specific compounds may be an effective approach for abrogating resistance to proteasome inhibition and effectively treating these hematologic cancers.
Table 1. Small molecule inhibitors of LonP1.
Non-peptide inhibitors of LonP1 have also been described. A boronic acid-based non-peptide inhibitor called compound 14 shows high affinity for LonP1 with an IC50 of 0.059 µM, with no cross-reactivity with the proteasome [Citation35] (). Whether this compound inhibits LonP1-mediated degradation of protein substrates in mitochondria has not been solidly demonstrated. Rather, compound 14 was shown to increase the levels of certain proteins co-immunoprecipitated with LonP1 in compound-treated cells [Citation35]. In addition, another study showed that non-peptide coumarinic derivatives compounds 4 to 8 inhibit recombinant LonP1 [Citation36] (). The ability to inhibit mitochondrial LonP1 in cells was not examined.
Additionally, two classes of plant-derived LonP1 inhibitors, Obtusilactone A and Sesamin, have been shown to block proteolysis by recombinant LonP1 with IC50 values of 34.1 μM and 19.9 µM, respectively, and in cultured cells they stabilize the reported endogenous protein substrate mitochondrial aconitase [Citation37]. Computer modeling using the crystal structure of the E. coli Lon protease domain suggest that these compounds interact with the proteolytic active site. These compounds were found to induce apoptosis in human lung cancer cells. However, further work is required to determine whether the inhibition of LonP1 by these compounds plays a role in promoting cancer cell death.
More recent work demonstrates that the synthetic triterpenoids CDDO and its derivatives inhibit LonP1 via a novel allosteric mechanism, blocking ATPase activity in a noncompetitive manner [Citation34]. ATP hydrolysis is required for the degradation of folded and partially folded protein substrates and for the chaperone function of LonP1 [Citation7,Citation9,Citation40]. By contrast, the cleavage of small peptides does not require ATP hydrolysis, however, peptidase activity is stimulated by binding of ATP or non-hydrolyzable ATP analogs [Citation41]. CDDO derivatives inhibit the ATP-dependent protease activity of purified LonP1 in the micromolar range- CDDO (IC50 = 13 µM), CDDO-Me (methyl) (IC50 = 1.9 µM), and CDDO-Im (imidazole) (IC50 = 2 µM). Notably, CDDO-Me selectively inhibits the ATPase activity of purified LonP1 but not that of the purified 26S proteasome. The ability of CDDO derivatives to inhibit LonP1 within the mitochondrial matrix is supported by data showing the co-localization of biotinylated CDDO with the organelle stain Mitotracker Green [Citation42] and, moreover, data showing that these compounds inhibit LonP1-dependent degradation of its endogenous protein substrates [Citation6,Citation34]. Importantly, it must be noted that CDDO derivatives have multiple cellular targets and are not LonP1-specific. Their direct binding sites have been identified for Keap1 [Citation43], IκB kinase beta (IKK-β) [Citation44], Jak1 and Stat3 [Citation45]. Nevertheless, CDDO and its derivatives offer opportunities for identifying the specific allosteric binding pocket(s) within LonP1 and for the discovery of specific, high affinity compounds effectively inhibiting this key AAA+ protease.
6. Expert opinion
Allosteric modulators provide several physicochemical advantages compared to compounds that bind to the active site of a target protein. These modulators are typically lipophilic, enabling them to establish stronger hydrophobic interactions with higher affinity and selectivity that can reduce off-target effects. Additionally, these compounds tend to exhibit a high degree of rigidity, which confers greater overall stability. The discovery of specific, high-affinity allosteric inhibitors of the LonP1 ATPase holds promise for disabling the protease and chaperone-like functions of this stress response protein, which is hijacked by cancer cells for their proliferative advantage.
The combinatorial inhibition of AAA+ proteases such as the 26S proteasome, LonP1 and ClpXP is a potentially efficacious and novel approach for anticancer therapeutics. Simultaneously inhibiting LonP1 and the proteasome could potentially overcome the partial resistance to proteasome inhibitors observed in multiple myeloma [Citation39]. Similarly, a combined strategy for inhibiting LonP1 and the mitochondrial ATP-dependent protease ClpXP may be efficacious in promoting cancer cell death as suggested by the finding that depleting the LONP1 and CLPP genes in prostate cancer cells synergistically attenuated cell growth and induced cell death [Citation26]. Small molecules inhibiting or hyperactivating ClpP have been identified, which selectively induce cancer cell death to a greater extent than normal cell death [Citation46], further supporting a combinatorial therapeutic approach. In the future, a more detailed and comprehensive mechanistic understanding of cancer cell reliance on mitochondrial AAA+ proteases like LonP1, ClpXP and related proteases is essential. This knowledge will inform the development of small molecules capable of disabling these specific functions, thereby facilitating the discovery of innovative anticancer therapeutics and approaches.
Article highlights
LonP1 has multifaceted functions as an ATP-dependent protease that degrades abnormal proteins and rate-limiting protein in mitochondrial metabolism, as an ATP-dependent chaperone promoting protein folding and assembly, and preventing aggregation, and as an evolutionarily conserved DNA-binding protein.
Upregulation of LonP1 in many cancer tissues and its involvement in cancer-associated pathways make it a promising new target for pharmacological intervention.
Allosteric inhibitors of LonP1 that block its ATPase activity provide an opportunity for identifying the allosteric compound-binding pocket(s) and the discovery of specific, high-affinity inhibitors.
The combination of LonP1 inhibitors and other cytotoxic agents may be a novel approach to overcome drug resistance in some cancers.
Abbreviations
| AAA+ | = | ATPases Associated with diverse cellular Activities |
| Akt | = | Protein Kinase B |
| CDDO | = | 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid |
| CDDO-Im | = | CDDO-imidazole |
| CDDO-Me | = | CDDO-methyl |
| COX | = | Cytochrome c oxidase complex |
| ETC | = | Electron transport chain |
| GTEx | = | Genotype-Tissue Expression |
| HIF-1α | = | Hypoxia-inducible factor-1 alpha |
| HPA | = | Human Protein Atlas |
| ISR | = | Integrated Stress Response |
| mtDNA | = | Mitochondrial DNA |
| OXPHOS | = | Oxidative phosphorylation |
| PDH | = | Pyruvate dehydrogenase |
| PDK1 and PDK4 | = | PDH kinases 1 and 4 |
| PDP | = | PDH phosphatase |
| ROS | = | Reactive oxygen species |
| UPRER | = | Unfolded Protein Response of the Endoplasmic Reticulum |
| UPRmt | = | Mitochondrial Unfolded Protein Response |
| TCGA | = | The Cancer Genome Atlas |
| TSVdb | = | TGCA Splicing Variants Database |
Declaration of interests
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Glynn SE. multifunctional Mitochondrial AAA proteases. Front Mol Biosci. 2017;4:34. doi: 10.3389/fmolb.2017.00034
- Quirós Pedro M, Español Y, Acín-Pérez R, et al. ATP-Dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8(2):542–556. doi: 10.1016/j.celrep.2014.06.018
- Gibellini L, De Gaetano A, Mandrioli M, et al. The biology of Lonp1: more than a mitochondrial protease. Int Rev Cell Mol Biol. 2020;354:1–61.
- Granot Z, Kobiler O, Melamed-Book N, et al. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by lon protease: the unexpected effect of proteasome inhibitors. Mol Endocrinol. 2007 Sep;21(9):2164–77.
- Tian Q, Li T, Hou W, et al. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J Biol Chem. 2011 Jul 29;286(30):26424–30. doi: 10.1074/jbc.M110.215772
- Lu B, Lee J, Nie X, et al. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ lon protease. Mol Cell. 2013 Jan 10;49(1):121–32. doi: 10.1016/j.molcel.2012.10.023
- Shin CS, Meng S, Garbis SD, et al. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat Commun. 2021 Jan 11;12(1):265. doi: 10.1038/s41467-020-20597-z
- Bezawork-Geleta A, Brodie EJ, Dougan DA, et al. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep. 2015 Dec 02;5(1):17397.
- Hori O, Ichinoda F, Tamatani T, et al. Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J Cell Bio. 2002 Jun 24;157(7):1151–60. doi: 10.1083/jcb.200108103
- Crewe C, Schafer C, Lee I, et al. Regulation of pyruvate dehydrogenase kinase 4 in the heart through degradation by the lon protease in response to mitochondrial substrate availability. J Biol Chem. 2017 Jan 6;292(1):305–312. doi: 10.1074/jbc.M116.754127
- Nimmo GAM, Venkatesh S, Pandey AK, et al. Bi-allelic mutations of LONP1 encoding the mitochondrial LonP1 protease cause pyruvate dehydrogenase deficiency and profound neurodegeneration with progressive cerebellar atrophy. Hum Mol Genet. 2019 Jan 15;28(2):290–306. doi: 10.1093/hmg/ddy351
- Haynes CM, Hekimi S, Driscoll M. Mitochondrial dysfunction, aging, and the mitochondrial unfolded protein response in caenorhabditis elegans. Genetics. 2022 Nov 30;222(4). doi: 10.1093/genetics/iyac160
- Lu B, Yadav S, Shah PG, et al. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J Biol Chem. 2007 Jun 15;282(24):17363–74. doi: 10.1074/jbc.M611540200
- Liu T, Lu B, Lee I, et al. DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J Biol Chem. 2004 Apr 2;279(14):13902–10. doi: 10.1074/jbc.M309642200
- Yang Q, Liu P, Anderson NS, et al. LONP-1 and ATFS-1 sustain deleterious heteroplasmy by promoting mtDNA replication in dysfunctional mitochondria. Nat Cell Biol. 2022 Feb;24(2):181–193.
- Strauss KA, Jinks RN, Puffenberger EG, et al. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ lon protease. Am J Hum Genet. 2015 Jan 8;96(1):121–35. doi: 10.1016/j.ajhg.2014.12.003
- Dikoglu E, Alfaiz A, Gorna M, et al. Mutations in LONP1, a mitochondrial matrix protease, cause CODAS syndrome. Am J Med Genet A. 2015 Jul;167(7):1501–9.
- Fukuda R, Zhang H, Kim JW, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007 Apr 6;129(1):111–22. doi: 10.1016/j.cell.2007.01.047
- Pryde KR, Taanman JW, Schapira AH. A LON-ClpP proteolytic axis degrades complex i to extinguish ros production in depolarized mitochondria. Cell Rep. 2016 Dec 6;17(10):2522–2531.
- Vaseva AV, Marchenko ND, Ji K, et al. p53 opens the mitochondrial permeability transition pore to trigger necrosis [research support, N.I.H., extramural]. Cell. 2012 Jun 22;149(7):1536–48. doi: 10.1016/j.cell.2012.05.014
- Sung YJ, Kao TY, Kuo CL, et al. Mitochondrial lon sequesters and stabilizes p53 in the matrix to restrain apoptosis under oxidative stress via its chaperone activity. Cell Death Dis. 2018 Jun 13;9(6):697. doi: 10.1038/s41419-018-0730-7
- Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013 Feb;1833(2):410–6. doi: 10.1016/j.bbamcr.2012.02.019
- Tian X, Zhang S, Zhou L, et al. Targeting the integrated stress response in cancer therapy. Front Pharmacol. 2021;12:747837. doi: 10.3389/fphar.2021.747837
- Bernstein SH, Venkatesh S, Li M, et al. The mitochondrial ATP-dependent lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood. 2012;119(14):3321–3329. doi: 10.1182/blood-2011-02-340075
- Goto M, Miwa H, Suganuma K, et al. Adaptation of leukemia cells to hypoxic condition through switching the energy metabolism or avoiding the oxidative stress. BMC Cancer. 2014 Feb 10;14(1):76. doi: 10.1186/1471-2407-14-76
- Lee YG, Kim HW, Nam Y, et al. LONP1 and ClpP cooperatively regulate mitochondrial proteostasis for cancer cell survival. Oncogenesis. 2021 Feb 26;10(2):18. doi: 10.1038/s41389-021-00306-1
- Ghosh JC, Seo JH, Agarwal E, et al. Akt phosphorylation of mitochondrial Lonp1 protease enables oxidative metabolism and advanced tumor traits. Oncogene. 2019 Oct;38(43):6926–6939.
- Uhlén M, Björling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol & Cell Proteomics. 2005 Dec 01;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200
- The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 10/21/23. [Internet]. 2023
- LonP1 transcriptomics - these data here are in whole or part based upon data generated by the TCGA research network [Internet]. 2023. Available from: https://www.cancer.gov/tcga
- Zanini G, Selleri V, De Gaetano A, et al. Differential expression of Lonp1 isoforms in cancer cells. Cells. 2022;11(23):3940. doi: 10.3390/cells11233940
- Park JE, Miller Z, Jun Y, et al. Next-generation proteasome inhibitors for cancer therapy. Transl Res. 2018 Aug;198:1–16.
- Frase H, Hudak J, Lee I. Identification of the proteasome inhibitor MG262 as a potent ATP-dependent inhibitor of the Salmonella enterica serovar typhimurium Lon protease. Biochemistry. 2006 Jul 01;45(27):8264–8274.
- Lee J, Pandey AK, Venkatesh S, et al. Inhibition of mitochondrial LonP1 protease by allosteric blockade of ATP binding and hydrolysis via CDDO and its derivatives. J Biol Chem. 2022 Mar 01;298(3):101719. doi: 10.1016/j.jbc.2022.101719
- Kingsley LJ, He X, McNeill M, et al. Structure-based design of selective LONP1 inhibitors for probing in vitro biology. J Med Chem. 2021 Apr 22;64(8):4857–4869. doi: 10.1021/acs.jmedchem.0c02152
- Bayot A, Basse N, Lee I, et al. Towards the control of intracellular protein turnover: mitochondrial lon protease inhibitors versus proteasome inhibitors. Biochimie. 2008 Feb 01;90(2):260–269. doi: 10.1016/j.biochi.2007.10.010
- Wang HM, Cheng KC, Lin CJ, et al. Obtusilactone a and (-)-sesamin induce apoptosis in human lung cancer cells by inhibiting mitochondrial lon protease and activating DNA damage checkpoints. Cancer Sci. 2010 Dec;101(12):2612–2620.
- Shin M, Watson ER, Song AS, et al. Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Nat Commun. 2021 May 28;12(1):3239. doi: 10.1038/s41467-021-23495-0
- Maneix L, Sweeney MA, Lee S, et al. The Mitochondrial Protease LonP1 Promotes Proteasome Inhibitor Resistance in Multiple Myeloma. Cancers (Basel). 2021 Feb 17;13(4):843. doi: 10.3390/cancers13040843
- Rep M, van Dijl JM, Suda K, et al. Promotion of mitochondrial membrane complex assembly by a proteolytically inactive yeast lon. Science. 1996 Oct 4;274(5284):103–6. doi: 10.1126/science.274.5284.103
- Lee I, Berdis AJ, Suzuki CK. Recent developments in the mechanistic enzymology of the ATP-dependent lon protease from Escherichia coli: highlights from kinetic studies. Mol Biosyst. 2006 Oct;2(10):477–83. doi: 10.1039/b609936j
- Samudio I, Konopleva M, Pelicano H, et al. A novel mechanism of action of methyl-2-cyano-3,12 dioxoolean-1,9 diene-28-oate: direct permeabilization of the inner mitochondrial membrane to inhibit electron transport and induce apoptosis. Mol Pharmacol. 2006 Apr;69(4):1182–93.
- Cleasby A, Yon J, Day PJ, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6):e98896. doi: 10.1371/journal.pone.0098896
- Ahmad R, Raina D, Meyer C, et al. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on cys-179. J Biol Chem. 2006 Nov 24;281(47):35764–35769. doi: 10.1074/jbc.M607160200
- Ahmad R, Raina D, Meyer C, et al. Triterpenoid CDDO-methyl ester inhibits the Janus-activated kinase-1 (JAK1)–>signal transducer and activator of transcription-3 (STAT3) pathway by direct inhibition of JAK1 and STAT3. Cancer Res. 2008 Apr 15;68(8):2920–2926.
- Nouri K, Feng Y, Schimmer AD. Mitochondrial ClpP serine protease-biological function and emerging target for cancer therapy. Cell Death Dis. 2020 Oct 9;11(10):841.

