ABSTRACT
Introduction
Immunotherapies have revolutionized cancer treatment, but often fail to produce desirable therapeutic outcomes in all patients. Due to the inter-patient heterogeneity and complexity of the tumor microenvironment, personalized treatment approaches are gaining demand. Researchers have long been using a range of in-vitro assays including 2D models, organoid co-cultures, and cancer-on-a-chip platforms for cancer drug screening. A comparative analysis of these assays with their suitability, high-throughput capacity, and clinical translatability is required for optimal translational use.
Areas covered
The review summarized in-vitro platforms with their comparative advantages and limitations including construction strategies, and translational potential for immuno-oncology drug efficacy assessment. We also discussed end-point analysis strategies so that researchers can contextualize their usefulness and optimally design experiments for personalized immunotherapy efficacy prediction.
Expert opinion
Researchers developed several in-vitro platforms that can provide information on personalized immunotherapy efficacy from different angles. Image-based assays are undoubtedly more suitable to gather a wide range of information including cellular morphology and phenotypical behaviors but need significant improvement to overcome issues including background noise, sample preparation difficulty, and long duration of experiment. More studies and clinical trials are needed to resolve these issues and validate the assays before they can be used in real-life scenarios.
1. Introduction
Harnessing the immune system to fight cancer has revolutionized cancer treatment providing a better outcome compared to conventional systemic therapies [Citation1]. One of the 14 hallmarks of cancer is immune evasion or evading immune destruction where cancer cells escape immune surveillance to survive and proliferate [Citation2]. Cancer immunotherapies target one or multiple pathways of communication between the tumor microenvironment and the immune system using biological components including specialized antibodies, modified immune cells, etc. This enhances the immune system’s ability to better detect and eliminate cancer cells with more precision and fewer system effects. Several immunotherapy drugs especially Immune Checkpoint Blockers (ICBs) are now clinically approved as the standard of care treatment for different advanced tumors including advanced melanoma and non-small cell lung cancer [Citation3,Citation4].
Despite improving clinical outcomes and response rates a large proportion of patients remain refractory to ICB treatments and suffer from adverse effects [Citation5]. In the case of metastatic advanced melanoma, two of the ICB classes are used in clinical settings which are PD-1 inhibitors (Pembrolizumab and Nivolumab) and CTLA-4 inhibitors (Ipilimumab). These drugs in combination (Nivolumab and Ipilimumab) have improved the treatment outcome for advanced melanoma patients with an overall survival rate of 49% and progression-free survival rate of 34% at 6.5 years but a large proportion of patients still remain refractory to the treatment [Citation6]. A systematic review identified that 74% of the patients treated with PD-1 inhibitors (PD-1i) have suffered from immune-related adverse effects and for CTLA-4 inhibitors (CTLA-4i) the rate was 89% [Citation7].
Due to the adverse effects and high non-response rate, it is often difficult to make optimal clinical decisions for patients considering both survival and quality of life parameters. Researchers have successfully tried ex-vivo drug efficacy assessment using multiple in-vitro assays for several hematological and solid cancers. These assays have also been useful in high-throughput screening and identification of novel immuno-oncology drugs. In this review, we gather information and compare multiple in-vitro assays for immuno-oncology efficacy assessment with their advantages and limitations.
For this narrative review, we have conducted electronic searches in Google Scholar, PubMed, and Web of Science. Searches were also conducted on the ‘Clinicaltrials.gov’ website to identify clinical trials on the topic. Relevant search keywords were used including ‘immunotherapy,’ ‘onco-immune,’ ‘efficacy,’ ‘organoid,’ ‘spheroid,’ ‘coculture,’ ‘ex-vivo,’ ‘in-vitro,’ ‘scaffold based,’ ‘scaffold free,’ ‘microenvironment,’ ‘immune interaction,’ ‘PDE,’ ‘organ on a chip,’ ‘3D model,’ ‘immunotherapy drug screening,’ and ‘immunotherapy efficacy prediction.’ The search results were filtered for the English language only. The last search was conducted on 16 February 2024. Publications were selected based on relevance (determined by the authors) and further searches were conducted to expand specific topics of interest. The most recent publication was considered where multiple publications were available.
2. Importance of pre-clinical drug efficacy assessment
Cancer is a complex heterogeneous condition and yet the treatment options are tumor-type-led in many cases, rather than patient-tailored. Thus, the effectiveness of these treatments in yielding desirable outcomes often varies and cannot always be predicted using traditional biomarkers. Moreover, due to the high plasticity of cancer cells, they rapidly develop multiple resistant subpopulations capable of surviving targeted inhibitors and immunotherapies, hence the disease frequently recurs through clonal expansion of resistant subpopulations despite excellent primary response [Citation8]. As discussed earlier, a large portion of patients (40–80% depending on cancer and therapy type) remain refractory to immunotherapies and may still suffer from adverse effects especially the responders [Citation7,Citation9]. So, more personalized treatment options including novel or repurposed drugs and combination therapies need to be identified to improve treatment outcomes in these cohorts of patients. The development of newer therapies addressing individual heterogeneity and the plasticity of the disease is a dire need.
Risk stratification of patients to improve treatment outcomes is another challenge due to the lack of reliable biomarkers. Researchers reported several pre-treatment biomarkers to predict response to immunotherapy in solid cancers but most of them have failed to be reliable enough to be clinically incorporated except for PD-L-1 expression, tumor mutational burden, and microsatellite instability [Citation10]. These biomarkers often fail to predict prognosis in the long term due to their instability over time and cancer heterogeneity as discussed earlier. Preclinical drug sensitivity testing could eliminate the need for biomarker validation and increase our understanding of the complex effect of genetic aberrations on individual patients. Patient-derived xenograft models are considered the gold standard for studying drug efficacy on tumors, but the process requires a significant amount of time with complex maintenance procedures that make it impractical for clinical use as a personalized treatment tool. Researchers have used many different clinically usable in-vitro models ranging from traditional cell culture to tumor-on-a-chip models that can be used to produce reliable and reproducible predictive results of personalized drug sensitivity within a period [Citation11].
3. In-vitro models for immunotherapy assessment
In-vitro efficacy assessment for immuno-oncology drugs is much more complex than other drugs such as chemotherapies or targeted inhibitors as it relies on direct and indirect assessment of immune cell functionality rather than cancer cells. As immunotherapies potentiate immune cells to target malignant cells, the efficacy assessment often needs to assess the complex spatial interactions between effector and target cells or measure biochemical crosstalk toward effective cell death. These can be achieved by mimicking the tumor microenvironment (TME) in-vitro, commonly using co-culture (culturing different cell types together) assays where immune cells and tumor cells are co-cultured within specially prepared cell culture systems [Citation12]. Multicellular 3D structures of tumor cells either in the form of spheroids or more specialized organoids are popular among researchers for co-culture and drug screening studies.
Many of the challenges often remain with generating these 3D structures. The most common challenge is ensuring the reproducibility and uniformity of spheroids. Multicellular tumor spheroid (MCTS) formation largely varies on cell type, formation is linked to various factors, such as cell type, culture technique, medium composition and volume, and cell density. These factors cause variability in MCTS formation, resulting in difficulties in reproducible spheroid formation. The advantages and limitations of different in vitro culture models are discussed below.
3.1. 2D models
2D cell culture models are simple and well-established for drug efficacy assessment. These can also be used as a co-culture system where both target (cancer cells) and effector cells (immune cells) are co-cultured in culture plates as a heterogeneous cell suspension without the need for generating complex 3D spheroids/organoids. The major advantages of 2D culture models are simplicity, low maintenance, easy to interpret, high throughput capability, and extensive use in high throughput screening that allowed the establishment of highly reproducible assays [Citation13,Citation14]. However, these models do not represent in-vivo cell-cell interactions as a large part of the cell surface remains inaccessible due to the attachment with the culture plate surface. As the adhered cells get flattened, the alteration in cellular morphology results in the alteration of gene expression, cell signaling, organization of intra-cellular organelles, polarity, and response to the external environment [Citation15–21]. Due to the lack of representation of the TME in 2D models, it is highly challenging to study cancer-immune interactions in response to therapeutics. Researchers have established a few models to reflect TME in 2D systems by co-culturing different cell types together (including cancer cells, antigen presenting cells, lymphocytes, fibroblasts, etc.) and allowing cancer-immune interactions which can be detected by different assays (immune cell activation, phagocytosis, cytotoxicity, image-based spatial interaction, etc.) [Citation22].
Much of these studies are based on established cell lines due to their known stability in different culture environments. Stock and colleagues conducted a comparative analysis of 2D and 3D models for drug discovery using cell lines (MCF7, LNCaP, NCI-H1437). They demonstrated the applicability of 2D co-culture platform in assessing tumor-stroma interactions. Using fluorescence image-based analysis they showed stroma derived growth benefit and possible drug resistance in MCF7 cells when co-cultured with human dermal fibroblasts [Citation23]. Mo and colleagues described another image based 2D co-culture platform where they screened inhibitor of apoptosis (IAP) antagonists as immune enhancers. They used colorectal cancer cell lines (SW48, LIM1215, and NCI-H838) and human Peripheral Blood Mononuclear Cells (PBMCs) as targets and effectors and their viability was measured by cellular fluorescence intensity and resazurin based biochemical assay [Citation24].
The example of using 2D co-culture platforms for ex-vivo immune-oncology drug efficacy prediction is scarce due to their limited capability in TME representation and supraphysiological access to the external environment in addition to the individual heterogeneity of patient samples [Citation25]. Pimentel and colleagues described an in-vitro 2D coculture model where they assessed the antitumor activity of irradiated and/or immunotherapy-treated (in-vivo) effector T-cell on in-vitro irradiated tumor cells using flow cytometry and luminescence-based cell viability assay. Although they used murine models and in-vivo treatment, they suggested the model could also be adapted to ex-vivo high-throughput immunotherapy screening using human patient tissue [Citation26].
3.2. 3D co-culture models
Compared to 2D, the 3D models are far more advanced and more closely mimic the TME as they can preserve cellular heterogeneity, polarization, morphology, and gene expression, and provide more surface area to study cell-cell interactions [Citation27]. The 2D monolayer cell cultures do not resemble tumor structure hence cell-cell interactions cannot be properly studied, even with primary culture models. Studying such interactions is valuable especially for immunotherapy screening as the immunotherapy drugs themselves may not exert cytotoxic effects on tumor cells but rather facilitate the immune system to kill them via changing cellular morphology and interaction. To overcome this problem many different techniques have been developed to allow the organization and growth of cells in a 3D spherical shape that more closely mimics the original tumor microenvironment [Citation28]. When such 3D structures are developed from patient-derived tumor samples, the process is commonly referred to as patient-derived organoids (PDOs). The terms spheroid and organoid are interchangeably used in literature to refer to compact 3D cellular aggregates often spherical in shape that structurally and sometimes functionally can mimic the original tissue it was derived from [Citation29]. To avoid confusion, we have replaced the term ‘organoid’ with ‘spheroid’ throughout the review.
Mäkelä and colleagues reported successful use of both 2D and 3D patient-derived culture models for personalized drug efficacy screening in a rare metastatic urachal carcinoma [Citation30]. One of the disadvantages of these models is the depletion of nonmalignant cells. This can be minimized by co-culturing the spheroids with peripheral blood-derived or other organ-derived immune and stromal cells that allow a continuous supply of such cells for maintaining the tumor microenvironment [Citation24,Citation31]. Such co-culture methods can also be used with cell lines giving rise to heterogeneous spheroids of cell lines closely resembling tumor microenvironment [Citation32].
One of the key challenges of using 3D co-culture models for drug efficacy assessment is to create and replicate uniformly shaped 3D structures from tumor cells/tissue for different cancer types. With the continuous evolution of 3D cell culture techniques, numerous types of 3D structures have been developed using different cell types. Each of these structures and techniques has its limitations and advantages (). These structures can be grown using single-cell suspensions from established cell lines (homogenous), patient-derived tissue dissociated cells (heterogenous), or tissue-derived stem cells that can functionally differentiate into miniature organ-like structures.
Table 1. Comparison of different 3D structural models for cancer research [30, 42, 65, 79, 80].
provides an overview of different 3D models. Among the different 3D structural models, MCTS is the simplest and most commonly used. MCTS are cellular aggregates that form compact spheroids either spontaneously or using external force (e.g. gravity, centrifuge, magnetic force) in low attachment culture conditions (). Single cell suspension from cell lines or dissociated patient tissue are cultured in low attachment plates or specially designed containers (spinner flask, bioreactor, microwell plates, etc.) that prevent cell adhesion on container surface and facilitate cell-cell adhesion leading to the tightly packed spheroid formation (). To facilitate MCTS formation for some cancer types, different media additives such as Methyl Cellulose can be used to increase the viscosity of media, thus facilitating the formation of cell aggregates [Citation50]. Sometimes artificial or animal-derived acellular scaffolds mimicking ECM are incorporated in culture conditions to facilitate MCTS formation. MCTS are generally easy to form within a short period of time (1–7 days), well characterized, high throughput capable, heterogenous, and represent in-vivo tumor characteristics making them a common choice for preclinical drug efficacy experiments [Citation29]. However, their uniformity in size and shape, response to environment, and metabolic activity often remain challenging which may cause a reproducibility issue. MCTS are excellent for the mass production of spheroids, enabling them to be used in drug screening studies, but only a limited number of cancer cells (e.g. MCF-7, HCT-116, RF-1. PANC-1, T47D) spontaneously form compact spheroids, while many others (e.g. SK-BR3, SW620, SNU-16, MIA PaCa-2, MDA-MB-231) form loose aggregates [Citation51].
Table 2. Comparison of spheroid forming techniques [Citation38,Citation40–49,Citation52,Citation62,Citation69,Citation95–100].
Tumorospheres are quickly formed stable compact spheroids but lack heterogeneous complexity as they are clonally expanded from single cancer stem cells. Dissociated cells are cultured in special ‘stem cell medium’ which generally lacks fetal bovine serum and is often supplemented by stem cell growth factors including epidermal growth factor, fibroblast growth factor, insulin, hydrocortisone, etc. Cells grown in low attachment plates at low seeding density using stem cell medium facilitate stem cell proliferation and deplete nonmalignant and differentiated cells giving rise to tumorospheres. They can be used to study cancer cell stemness but due to the lack of heterogeneous complexity, their use in drug screening may not correspond with clinical scenarios [Citation52]. Heterogenous spheroids can also be obtained from partially dissociated malignant tissue which are known as tissue-derived tumor spheroids (TDTS). Colospheres are one of the common examples of TDTS where finely cut tumor tissue fragments are minced for mechanical dissociation and cultured in culture flasks that give off circular compact spheroids within a day [Citation53]. In some cancer types culturing small pieces (300 to 800 µm) of tumor tissue fragments can give off compact spheroids similar to TDTS without any mechanical or enzymatic dissociation steps. These are commonly known as organotypic multicellular spheroids (OMS). Burgues and colleagues reported successful generation of bladder cancer OMS from biopsy tissue fragments within 2 days of culture in agarose coated 25 mm2 flask and harvested the spheroids by careful pipetting under a light microscope [Citation36]. Both TDTS and OMS are quick and easy to form but their initiation success rates are poor for different types of cancer. Generated spheroids are highly variable in size and shape and not suitable for high throughput screening due to a limited number of spheroid generation capacity [Citation29].
Researchers have also reported using patient derived explant (PDE) culture platform for ex-vivo drug screening including immuno-oncology drugs without generating spheroids. In PDE culture larger tissue fragments (1–2 mm) are directly cultured either submerged in media or keeping in partial contact with media through a membrane or matrix. As the tissue fragments are not dissociated, the TME remains intact preserving all the in-vivo tissue components. However, due to the size and compactness, they are low throughput, require larger tissue samples, not suitable for long term culture and very limited end-point analysis can be conducted [Citation37]. Vaobil and colleagues described using a PDE platform for clinically relevant ex-vivo immunotherapy response prediction in melanoma, non-small cell lung carcinoma, breast cancer, ovarian cancer, and renal cell carcinoma [Citation54].
With the incorporation of engineering techniques, researchers have developed microfluidics and 3D printing-based models that can be used for in-vitro immuno-oncology drug efficacy analysis. Organ-on-a-chip systems are gaining popularity as they allow precise manipulation of environmental components such as nutrients, oxygen, chemokines, etc. These chips are designed on a polydimethylsiloxane (PDMS) base with multiple compartments and channels to allow experimental freedom for fluidic and biochemical adjustments. Cui and colleagues conducted ex-vivo glioblastoma (GBM) culture on an organ-on-a-chip platform and identified that CSF-1 R inhibitor coadministration improves PD-1 inhibitor efficacy leading to GBM apoptosis. 3D-bioprinting is a more advanced technique where cellular and ECM components as well as biochemical factors can be precisely controlled and layered to form 3D structures that are structurally and morphologically more similar compared to self-assembled organoids [Citation55]. Heinrich and colleagues developed a 3D-bioprinting based GBM model where they co-cultured glioblastoma cells and GBM associated macrophages (GAMs) within bioprinted ‘mini-brains.’ They identified slower GBM cell growth and functional inhibition of GAMs in response to STAT6 and CSF-1 R inhibitors [Citation49]. Both models are robust in experimental freedom and suitable for ex-vivo drug screening. However, the requirement for external devices, complex maintenance procedures, and high cost compared to other methods makes their use challenging in both pre-clinical and clinical settings [Citation11,Citation56,Citation57].
A graphical summary of the models is displayed in below.
Figure 1. Different cell culture techniques to form spheroids from cancer cells. (a) Hanging drop: gravitational pull facilitates spheroid formation (b) suspension culture: plate surface modification prevents attachment and facilitates cell aggregation (c) high viscosity suspension: media additives (e.g. methyl cellulose) increase viscosity and facilitate cell aggregation (d) pellet culture: centrifugation assists spheroid formation (e) magnetic levitation: magnetized cells aggregates to form spheroid assisted by a magnetic field (f) spinner flask: continuous agitation prevents cell attachment and facilitate spheroid formation (g) microwell plate: micropatterned wells allow uniformly sized spheroid formation (h) hydrogel embedding: hydrogel containing extracellular matrix components (laminin, collagen, etc.) aid in spheroid formation (i) fibrous scaffold: decellularized or synthetic fibrous scaffold support spheroid formation in-vitro (j) bioprinting: cells within bio ink layered in computer-aided patterns supports the formation of 3d structures (k) tumor-on-a-chip: specially designed chips containing microwells and channels supports spheroid growth with dynamically controlled environment. The illustration is created with BioRender.com.
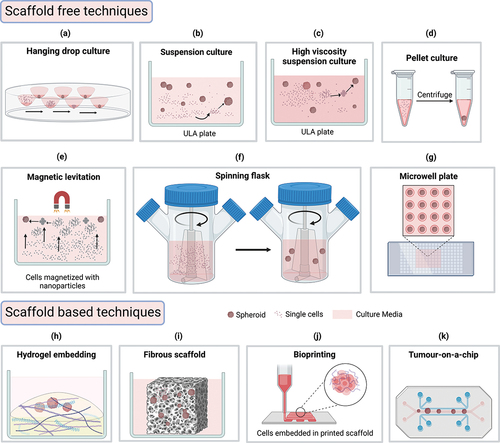
All the above-mentioned 3D structures can be generated using a wide range of techniques that researchers have developed over the years (). The techniques can be classified into two groups, Scaffold-free and Scaffold-based. The most used scaffold-free technique is suspension culture, also known as liquid overlay culture, which can mass-produce multicellular spheroids. The process involves using a low attachment surface that facilitates the spontaneous aggregation of cells into spherical 3D structures. Hydrophilic polymers such as Agarose, Poly 2-hydroxyethyl methacrylate (PolyHEMA), and Polyethylene glycol-block-poly propylene glycol-block-polyethylene glycol, are generally used to coat non-treated cell culture plates/dishes to produce a cell-repellent surface that forces the cells to float. Commercial ultra-low-attachment (ULA) plates/dishes are also available with low adhesion coating. This surface modification prevents cell adhesion to the inert surface and enables cell-cell interaction thus formation of spheroids occurs within a few days depending on the growth cycle of the cell types. Once the cells have formed spheroids of a desired size, they can be co-cultured with immune cells commonly isolated from blood as PBMCs to determine the therapeutic effects of drugs [Citation44,Citation58,Citation59]. This technique is frequently used due to its ease of initiation, maintenance, and harvest, low cost, and high throughput capability. However, this technique can only be used for a limited type of cancers, and it does not produce uniformly shaped and sized spheroids that may impact the reproducibility of end-point analysis [Citation52,Citation60,Citation61]. Tong and colleagues demonstrated the oncolytic potential of the Marab virus on ovarian cancer spheroids generated using suspension culture technique [Citation41].
Other scaffold free techniques involve using external aids to facilitate spheroid formation. In hanging drop culture single cell suspension drops are hung from a culture surface allowing gravity to force cells into forming compact aggregates. This is a quick and easy technique, but long-term culture is not possible as media changing in hanging drops is highly challenging. Another way of forcing cells into forming aggregates is increasing media viscosity by adding inert additives such as agarose or methylcellulose. Additives can be used in both hanging drop and suspension culture techniques, but additional clearing steps may be required during spheroid harvesting. Mukherjee and colleagues reported using 2% methylcellulose supplemented hanging drop method for mouse melanoma spheroid generation which they used in co-culture to demonstrate activated immune cell infiltration in co-culture [Citation42]. Spheroids can also be formed by centrifuging them to form pellets and culturing them as pellet culture in Eppendorf tubes. However, we could not find any study reporting its use in immune oncology experiments. External forces facilitating spheroid formation are also used in spinner flasks and bioreactors. Continuous agitation from spinning keeps cells in a floating state inside the spinner flask and horizontal rotation in the bioreactor creates microgravity forcing cells to form spheroids. Spinner flasks are suitable for mass production of spheroids but may not be cost effective and the shear forces generated from rotation may also damage cells as well as alter gene expression. Hence only cell lines that can withstand high shear force are suitable for this platform [Citation52,Citation62]. The shear force is comparatively low in bioreactors, but they are not cost-effective and need additional maintenance. Weiqi He and colleagues showed that HLA-E or NKG2A inhibition facilitates NK cell-based glioma cell line (U251) spheroids generated using the bioreactor method [Citation43]. Researchers have reported using magnetic force to facilitate spheroid generation where cells are magnetized using nanoparticles and cultured inside a magnetic field by placing magnets on top (levitation) or bottom (magnetic bioprinting) of the plate. The magnetic field forces cells to aggregate within a few hours thus initiating spheroid formation. While this is a quick and easy technique, the challenges remain in optimizing magnetic nanoparticle concentration and incubation duration for individual cell types [Citation63]. Ferreira and colleagues reported using magnetic levitation technique to generate saliva secreting spheroids from primary cell culture [Citation64].
All the above-mentioned techniques have a common limitation that they cannot generate uniformly shaped and sized spheroids which may be required to ensure reproducibility of experimental results. This challenge can be overcome by culturing cells in low attachment round bottom microwell plates where culture plates are patterned into tiny microwells allowing cells to grow within a limited amount of space. The drawbacks of this method are, this is not suitable for long term culture due to space restriction, and it requires special microwell patterning which are available commercially or can be generated by agarose molding using 3D printed PDMS stamps. Courau and colleagues identified the therapeutic efficacy of anti-MICA/B and anti-NKG2A facilitating NK cell mediated destruction of colorectal carcinoma spheroids generated from microwell plate culture [Citation44].
Many of the scaffold-free techniques such as suspension, hanging drop, pellet, microwell, and magnetic culture techniques are inexpensive and easy to use. However, these techniques are suitable for a limited number of cancer cell types as they lack an ECM environment which is a crucial part of TME and essential for many different cellular interactions including providing support for 3D structure formation in-vivo. To overcome this problem, several scaffold-based techniques have been developed that can closely mimic the ECM environment with the appropriate stiffness and structural stability required for specific tumor types. The in-vivo ECM components contain fibrous proteins (e.g. collagen, laminin, fibronectin, elastin), proteoglycans, and polysaccharides that provide matrix structure for cellular communication and organization. Commercial hydrogel solutions are available that are rich in these proteins and have been successfully used by researchers for 3D culture experiments. Hydrogel embedding is one of the widely used scaffold-based techniques where cells are embedded in ECM mimicking hydrogel rich in structural proteins and overlayed by liquid media. The natural proteins in hydrogel provide cell binding ligands and structural stiffness for spheroid formation from cells that may not form compact 3D shapes in scaffold free platforms. While this model provides a more realistic representation of TME, separating spheroids from the matrix can be difficult and also reported to have a batch-to-batch inconsistency [Citation52,Citation65]. Ou and colleagues demonstrated the personalized efficacy of anti-PD-1 antibodies on patient derived melanoma spheroids generated using hydrogel embedding technique. They directly added isolated tumor infiltrating lymphocytes on top of the hydrogel embedded spheroids and assessed their migration rate and infiltration using confocal microscopy [Citation45]. Decellularized or synthetic fibrous non-hydrogel scaffolds are also available that can provide structural support for spheroid formation. Cell removing agents are applied to healthy or malignant tissue to obtain the matrix which has the closest similarity to native ECM, but these matrices demand additional tissue extraction and can have a batch-to-batch inconsistency along with lower architectural control. Synthetic fibrous scaffolds do not have these limitations, but they are still under development to be used widely. Both synthetic and decellularized scaffolds have transparency issues making them unsuitable for image based co-culture experiments [Citation52,Citation65]. Wallstabe and colleagues reported receptor tyrosine kinase-like orphan receptor 1-specific CAR T cells exhibit antitumor effect when co-cultured with A549 and MDA-MB-231 spheroids grown using decellularized scaffold [Citation46].
The hydrogel embedding technique is less suitable for automation due to its gel like structure and requires careful, fast, and laborious handling. This can be resolved by using the 3D bio-printing technique described earlier. Layer wise deposition of cellular and biochemical materials allows more spatial control and biomimetic tissue architecture with higher reproducibility [Citation66]. Chip based (organ-on-a-chip) microfluidic culture systems are preferable for dynamic culture conditions which may require for long term culture with a better control of the environment. These devices allow both hydrogel based and scaffold free culture models to study migration, drug sensitivity, and immune infiltration effects [Citation52]. The limitations of these systems are that they require special handling and are less cost-effective. Mu and colleagues used a circulation system incorporated tumor-on-a-chip platform to demonstrate spatial infiltration of CD8+ T cells into hydrogel embedded ex-vivo tumor culture in response to combination immunotherapy [Citation48].
Finally, another method of 3D co-culture is air-liquid interface (ALI) where hydrogel embedded spheroids or minutely minced tissue pieces are partially kept in contact with culture media using trans-well inserts leaving the apical part of the tissue exposed to air for better oxygen access. Neal and colleagues used this technique to propagate patient derived spheroids and co-cultured them with TILs to assess the cytotoxic effect in response to PD-1/PD-L1 blockade [Citation47]. The comparative advantages and limitations of both scaffold-free and scaffold-based techniques are discussed in .
4. Endpoint analysis for immunotherapy efficacy prediction
For drug screening experiments, researchers commonly use viability/proliferation assays on drug-treated cells either by direct cell counting using fluorescence microscopy or indirect viability assessment by enzymatic assays, e.g. ATP-based bioluminescence assay. Some other methods of viability/proliferation assessment include flow cytometry and cytokine assessment. From these assays, variations of response over a range of drug concentrations compared to controls are assessed and can be used to generate multiple drug response metrics to statistically quantify these variations. The commonly used metrics are IC50 (the drug concentration at which cell population is halved compared to control), Emax (drug concentration at which the maximum effect of the drug is achieved), EC50 (drug concentration at which half of Emax is achieved), GR50 (drug concentration at which cell growth rate is 50% compared to control) and AUC (the cumulative effect of drug determined by the area under the drug dose-response curve), and DSS (Drug Sensitivity Score, based on closed-form integration of the area under the estimated dose-response curve) [Citation52,Citation67,Citation68].
While all these methods are well established and have been repeatedly used in numerous drug screening experiments on 2D platforms, their use in 3D co-culture models could be challenging, especially for immunotherapy drug screening. As 3D structures are more complex than 2D monolayer cultures, standard end-point assays can be adapted in addition to 3D model-specific assays for 3D drug screening experiments [Citation52]. Much of these assays depend on spheroid characterization by measuring shape, size, compactness, necrosis, and functional activity [Citation51]. Here we discuss some of the endpoint analyses that researchers have used or can be used for immuno-oncology drug efficacy assessment ().
Figure 2. Multiple methods of ICB efficacy assessment from spheroid-PBMC co-culture. (a) Cocultured spheroids are harvested and dissociated for flow cytometric analysis of T-Cells to compare activated T-cell population and tumor cell population levels (b) spheroids are stained with live-dead fluorescent staining and imaged to compare the proportions of dead cells between treatment and control (c) co-cultured spheroids are fixed and fluorescently labeled for imaging analysis where the spatial distribution of immune cells is compared (d) co-cultured spheroids are treated with ATP based bioluminescence assay for viability analysis (e) co-cultured well substrates are analyzed by ELISA method to compare levels of CD8+ activation enzymes e.g. Granzyme B. The illustration is created with BioRender.com.
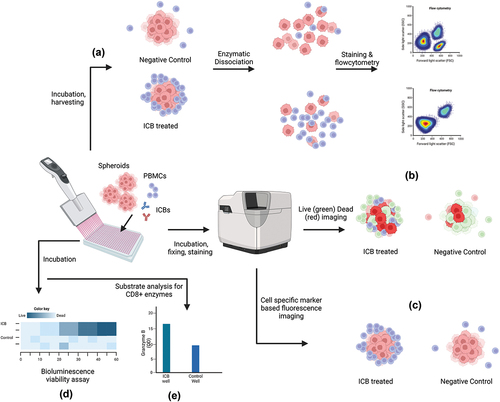
4.1. Metabolic viability/cytotoxicity assays
Several bioluminescence assays () have been adapted for 3D systems, which quantitatively measure luminescence signals from metabolic reactions in living/dying cells such as ATP-mediated luciferase reaction in living cells, mitochondrial dehydrogenase mediated WST-1 to formazan reaction in live cells, Caspase 3/7 mediate luciferase reaction in apoptotic cells, etc [Citation39,Citation52,Citation69].
Särchen and colleagues reported that BH3 mimetics can potentiate NK cell-based immunotherapy in Pediatric Rhabdomyosarcoma (RMS) as they have successfully demonstrated drug efficacy in RMS spheroid-coculture model using ATP based 3D cell viability assay [Citation70].
4.2. Enzymatic/cytokine quantification assays
In most cases, immunotherapies potentiate immune cells to kill tumor cells via receptor recognition followed by enzymatic degradation. Activated immune cells (e.g. Tumor Infiltrating Lymphocytes, NK cells) release cytokines (e.g. Granzyme B, IFNγ, Perforin-1) in response to tumor antigens and the level of cytokine can be measured in co-culture systems to assess immunotherapy efficacy, especially for ICBs ().
Wan and colleagues measured IFNγ levels in tumor media where High-Grade Serous Ovarian Cancer spheroids were co-cultured with immune cells. Compared with untreated controls, the researchers found significant elevations of IFNγ in the tumor media where spheroid cocultures were treated with PD-1/PD-L1/Bispecific antibodies [Citation71].
Meng and colleagues also used ELISA-based IFNγ and Granzyme B quantification in pancreatic tumor spheroid coculture supernatants to determine T-cell activation. They have used autologous PBMCs which were primed by co-culturing with tumor spheroids, which were subsequently used for immune checkpoint blockade efficacy assessment. The co-culture supernatant analysis revealed the highest IFN-γ production in response to NKG2A blockade compared to other ICBs and control [Citation72].
4.3. mRNA expression assay
Immune cell activity in response to tumor antigens and immunotherapy treatment can also be determined by mRNA expression analysis. Up or down-regulation of immune cell activation mRNA markers (e.g. IFNG, GZMB, GZMK, PRF1) can provide useful information about the alteration of tumor immune cell function in response to immunotherapy.
Neal and colleagues reported anti-PD-1 and anti-PD-L1 efficacy on heterogeneous spheroids propagated from > 100 patient/xenograft tumor samples using the ALI method. Upon qRT-PCR analysis of co-cultured T-cells, IFNG, GZMB, and PRF1 mRNA expression was significantly higher in anti-PD-1/PD-L-1 treated groups compared to the controls in their study [Citation47].
4.4. Flow cytometry assays
This endpoint analysis commonly involves dissociation of co-cultured tumor spheroids into single-cell states and counting cells of interest to compare treatment and control groups (). Decreased tumor cell number (expressing specific tumor antigen markers) or expansion of T-cell population (expressing T-cell activation markers) in dissociated co-cultured models can indicate the therapeutic efficacy of immuno-oncology drugs when compared with control. Courau and colleagues analyzed T and NK cell infiltration in colorectal carcinoma spheroid (HT29) using flow cytometry. They observed higher spheroid infiltration of immune cells in the presence of IL-15 as well as a higher proportion of T cell subsets expressing activation markers CD45RO, CD25, and CD107a [Citation44].
Flow Cytometry can also be used to quantify T-cell activation chemokines (IL-2, IL-4, TNF-α) in culture supernatants as described by Collins and colleagues. Briefly, specially designed beads with cytokine capture antibody along with a detection reagent (fluorophore-conjugated antibody) are incubated with cell culture supernatant that forms capture bead-analyte-detection reagent sandwich complex. This can be detected using flow cytometry and the analytes can be quantified by comparing with standards. Zhou and colleagues identified a significantly increased concentration of chemokines (CXCL10, CXCL9, CXCL8, CCL2, and CCL5) in HGC37 MCTS-PBMC co-culture in the presence of PD-1 blockers. They also identified a higher proportion of dead cells within the co-cultured spheroids (from fluorescence confocal imaging) in the presence of PD-1 blocker which correlated with the increased supernatant chemokine and suggests a possible contribution of chemokines in tumor cell killing by recruiting activated immune cells inside the spheroids in presence of PD-1 blocker [Citation73].
4.5. Images based assays
While most of the above-mentioned assays indirectly measure immunotherapy-assisted cytotoxicity or immune cell activity, image-based assays allow more direct spheroid characterization allowing more sensitive and diverse functional assessment. Microscopy-based techniques are more sensitive than other assays as they do not require spheroid dissociation for end-point analysis. Imaging can allow multiple endpoint assessments such as spheroid morphology characterization, viability assessment by fluorescent live/dead staining, the spatial distribution of immune cells surrounding spheroids to measure immune cell infiltration, etc. [Citation52,Citation74]. ().
The simplest of the imaging techniques is brightfield microscopy which provides information on tumor size and shape and can be used to identify immune cell invasion. Courau and colleagues reported brightfield microscopy can be used to determine the volumetric reduction of human colorectal tumor spheroids (HT29 cell line) when co-cultured with activated T & NK cells with or without IL-15 compared to controls [Citation44]. More complex imaging systems including SEM (Scanning Electron Microscopy), TEM (Transmission electron microscopy) and AFM (Atomic Force Microscopy) can assess fine and ultrastructural measures on spheroid surface or lumen (based on the 3D structure). Kast et al. reported a 3D coculture model for immunotherapy response assessment in pancreatic cancer, where they used SEM and AFM for spheroid morphology assessment including spheroid crowding density, ECM deposition, and matrix stiffness [Citation75].
Fluorescence microscopy techniques are more popular in drug efficacy assessment experiments as they provide viability as well as multiple marker expression information on different cell types allowing differential analysis in a mixed cell population. While this technique has been used widely in monolayer cultures, their use in 3D co-culture models is quite challenging due to factors including, low light penetrance, low marker penetrance, high light scattering from neighboring cells, autofluorescence, and high background [Citation52,Citation74]. Conventional microscopy approaches acquire images from a single plane which often fails to gather enough information from a 3D object. This can be solved by taking a series of equally distant XY images on the Z-axis (Z-stack) and processing them into a single image by overlapping [Citation76,Citation77]. For image acquisition, widefield microscopes are faster enabling it for high-throughput experiments but often it requires rigorous post-processing as they use LED lights which scatter more compared to laser excitation. On the other hand, confocal microscopes use laser excitation yielding a more detailed, clearer image with higher resolution, but it takes a longer time reducing its suitability for high-throughput use. Researchers have also reported the use of light sheet fluorescence microscopy (LSFM) which illuminates fluorophores on a single plane reducing out-of-focus signals from neighboring cells [Citation78,Citation79].
For fluorescence imaging-based analysis, different types of fluorophores can be used. One of the common practices is to fix and permeabilize spheroids and label them with specific primary antibodies that can be selective for specific cell types (e.g. tumor cell marker, immune cell marker) followed by the addition of secondary antibodies attached with fluorophores. These antibodies can also be directly labeled where the primary antibody is conjugated with a fluorophore. These fluorophores can be excited with a specific light source and imaged with a fluorescence microscope. The problem with this technique is that it does not allow live cell imaging and marker penetration can be low depending on spheroid size and density [Citation52]. To overcome this problem, different cell populations can be prelabelled with different nontoxic cell tracer markers such as Carboxyfluorescein Diacetate Succinimidyl Ester (CSFE) or 5-(6)-(((4-Chloromethyl)Benzoyl)Amino)-Tetramethylrhodamine (CMTMR) before co-culture and then traced within the spheroid to determine the population ratio for measuring infiltration [Citation80]. This technique uses live cells and time-lapse imaging, but the markers lose their fluorescence intensity as the cells undergo cell division. Alternatively live-dead cell markers such as Calcein Acetoxymethyl (Calcein AM) which emits green fluorescence inside live cells and Ethidium Homodimer-1 (EthD-1) or Propidium Iodide (PI) which emits red signals when bound with DNA inside the membrane-damaged dead cells. This technique can be used to determine live/dead cell proportion in response to treatment [Citation74].
A recent study reported the use of multiple fluorescence staining techniques and confocal microscopy to identify drug response on a patient-derived melanoma-TIL (tumor-infiltrating lymphocyte) 3D co-culture model. They reported a higher infiltration of CD3+ and CD8+ T cells within the spheroids in the presence of anti-PD-1 or anti-PD-1+IL2 treatment compared to control. They also observed a significant increase of dead cells within the spheroids in treatment groups using live-dead staining [Citation45].
Zhang and colleagues reported higher infiltration of B7-H3 CAR T cells inside HCT116 spheroids in the presence of a small molecule hedgehog inhibitor where cells were stained with Calcein/PI and CSFE and images were taken with a confocal microscope [Citation81].
5. Clinical readiness of in-vitro assays
We have discussed a substantial number of in-vitro assays that researchers have used successfully to assess immune-oncology efficacy on multiple cancer types. However, translating the assays into clinical practice is much more challenging. In clinical scenarios, we need to consider both inter-patient and intra-patient tumor heterogeneity that can alter cellular behavior in-vivo and in-vitro. As discussed earlier, different cancer types require different culture environments, yet established cell lines can be easily adapted to a wide range of immunotherapy efficacy assessment models (both 2D and 3D). Patient derived primary cells are extremely variable and may not behave similarly as cell lines. For example, cell lines can withstand shear force in a spinner flask and mass produce spheroids, but they are not suitable for primary cells [Citation52]. Scaffold free assays are easy to handle but they lack TME representation hence the screening results may not reflect clinical outcomes (). For the same reason, 2D co-culture platforms are also less reliable for clinical use. The PDE and ALI models preserve original tissue architecture but require a large amount of tissue samples which may not always be possible in clinical settings. Where tissue samples are limited and TME representation is required, hydrogel-based techniques can be used if reproducibility can be ensured through proper optimization. Spheroid uniformity issue can be resolved by using hydrogel embedding within micro-well plates. Microfluidic devices and 3D bio-printing techniques can be clinically useful as they provide better environmental control and incorporate dynamic culture conditions similar to in-vivo environments, but they are still under development and need rigorous testing to be optimized before clinical use [Citation52]. Researchers reported a degree of variability in 3D structure formation and end-point assay duration for different cancer cell lines as well as primary tumor cells which nay require up to 30 days [Citation29,Citation45,Citation55,Citation82–84]. This duration is too long to be useful in clinical settings as the clinicians often need to start treatment within a few days of surgery or biopsy especially for aggressive fast-growing cancers. To the best of our knowledge there are no published or ongoing clinical trials on personalized efficacy prediction of onco-immune drugs which are clinically relevant. We have found one multicenter phase II trial (RAMONA trial, NCT03416244) exploring clinical efficacy of nivolumab and ipilimumab as second line treatment in elderly advanced esophageal squamous cell carcinoma patients. They have included a translational sidearm in the study where they aim to identify novel biomarkers for checkpoint inhibition by using patient derived organoids at different time points of the study [Citation85].
6. Expert opinion
With the rapid development of immunotherapies for cancer treatment, the demand for suitable preclinical drug screening assays with higher predictive value is increasing day by day. The current preclinical drug screening gold standard animal models have served the purpose well, but the associated challenges including the duration of the assay, and scientific commitment to reducing animal use in research have made the models less desirable. On the other hand, high-throughput in-vitro models are gaining popularity due to their robust screening power of assessing large drug libraries within a short duration. However, multiple challenges of in-vitro models (including ex-vivo) remain along parameters including technical ease, reproducibility, resource needs, high-throughput capability, and tumor microenvironment (TME) mimicry [Citation51,Citation76]. Researchers have developed an array of techniques addressing these issues, but high-throughput, easier, and reproducible models lack proper TME and vice versa. For example, tumor-on-a-chip models are robust in mimicking TME including dynamic nutrient flow. Mu and colleagues demonstrated tumor-on-a-chip model can maintain nutrient and hypoxia gradients as well as cellular compositions similar to an in-vivo environment, but it is not suitable for high-throughput screening with a large number of drugs [Citation48]. On the other hand, scaffold-free MTCS models can quickly generate a large number of spheroids and their size and shape uniformity can be ensured using micropatterned wells allowing high-throughput capability, but lack of ECM environment doesn’t reflect in-vivo conditions hence less reliable for drug response analysis [Citation52]. For similar reasons, commonly used monolayer culture models lack in-vivo tissue architecture and fail to provide information including cellular morphology, complex cell-cell interactions in 3D, and spatial distribution, thus may not reliably predict therapeutic response in clinical settings. Although, several studies on hematological cancers have successfully demonstrated reproducible ex-vivo drug screening assays using 2D co-culture techniques as their in-vivo environment is less complicated to reconstitute in-vitro compared to solid tumors. Snijder and colleagues reported in their first-of-its-kind trial that image-based high-throughput ex-vivo screening on monolayer culture can guide treatment decisions with better outcomes compared to standard-of-care practice for multiple hematological malignancies [Citation86]. Recently they also reported an ex-vivo model for immunotherapy efficacy prediction in multiple myeloma [Citation87]. As reported in this article, several studies used enzymatic or flow cytometry-based end-point analysis to determine immunotherapy on ex-vivo culture models, but these assays destruct cell morphology and arrangement which makes the assays vulnerable to confounding and reliability. Researchers also reported both phenotypic and genotypic changes in monolayer culture over time which makes it even less reliable for complex immune interaction analysis required to determine immunotherapy efficacy [Citation88,Citation89]. Li and colleagues reported the use of both monolayer and 3D co-culture methods and multiple endpoint assays including luciferase assay, cytokine assay, and flow cytometric analysis of dissociated cells to determine the effect of tumor-associated macrophage modulation [Citation90].
Studies reported image-based 2D and 3D assays that can yield a wide range of information including morphology, cell death, cell population proportion, cell invasion, spatial interaction, immune cell activation, etc. which can complement each other to predict immuno-oncology drug efficacy with a much higher confidence. The key limitations of these assays are background signals from light scattering and high image acquisition time which is unsuitable for high-throughput screening. Ou and colleagues, in their recent paper, reported ECM-embedded confocal imaging-based ICB efficacy assessment on patient-derived melanoma spheroids. Their assays included cell type specific immunofluorescence staining to identify cell population diversity, live-dead staining to determine immunotherapy effectiveness, and cell tracer markers to detect immune infiltration rate on live cell imaging [Citation45].
Undoubtedly, the in-vitro cell culture-based immuno-oncology drug efficacy assessment has drawn the interest of researchers leading to the development of a variety of models and end-point assays. As useful as the models are, they are often limited to specific cancer or assay types yielding a limited number of information and requiring special considerations often failing to be reproducible, reliable, and high throughput. We are still in need of robust high-throughput assays that can simultaneously solve these issues which also need to be clinically validated through large-scale trials before it can be practically used for effective treatment decision making. So far, we have observed that multiple in-vitro assays are needed to complement each other to address complex immune interaction-related information for immunotherapy efficacy assessment in different cancers. Studies have reported the use of multiple assays to demonstrate drug efficacy in either cell line derived or patient-derived solid tumor organoids and a few of them have collated their results with clinical outcomes as well [Citation91–94]. However large-scale studies focusing on immunotherapies, especially clinically used or potential experimental ICBs are still lacking. We think imaging-based ex-vivo efficacy models are the way forward for personalized efficacy prediction of immuno-oncology drugs to be used in clinical settings or identifying novel agents or combination options.
Article highlights
Both 2D and 3D culture methods are available to model tumor immunity for immunotherapy efficacy prediction.
Patient-derived 3D organoid models comprising extracellular matrix (ECM) and heterogeneous cell populations similar to the original tumor can closely mimic in vivo tumor microenvironment and are more reliable for drug efficacy prediction.
Exogenous immune cells from peripheral blood or expanded tumor-infiltrating lymphocytes can be used in co-culture models for efficacy prediction of immunotherapy drugs.
Image-based end-point analyses are best suited to explain tumor-immune cell interaction within reconstituted in vitro models as well as analyze cell viability, immune infiltration, and activation effects of immuno-oncology drugs.
Complex models have reproducibility issues whereas commonly used 2D and less complex 3D models are unreliable in explaining tumor-immune interaction upon drug exposure.
Large-scale trials are needed for clinical validation of the successful models.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Declaration of interests
MM Rahman’s PhD studentship is supported by the Commonwealth Scholarship Commission, UK. G Wells is one of the investigators in the EVIDENT trial (NCT05231655). JK Rantala is the founder and owns shares in Misvik Biology Oy. T Helleday is the founder, on the board of directors and holds shares in One-Carbon and Oxcia and is one of the investigators on the EVIDENT trial (NCT05231655). SJ Danson is the principal investigator on the EVIDENT trial (NCT05231655). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Acknowledgments
We would like to thank H Gagg and S Hansford from the Sheffield Ex-vivo group and F Howard from the Nanobug Oncology Sheffield group for their insightful suggestions. The figures were created with Biorender.com.
Additional information
Funding
References
- Esfahani K, Roudaia L, Buhlaiga N, et al. A review of cancer immunotherapy: from the past, to the present, to the future. Curr Oncol [Internet]. 2020. [cited 2023 Nov 3];27(12):S87–97. Available from: /pmc/articles/PMC7194005/. doi: 10.3747/co.27.5223
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov [Internet]. 2022 [cited 2022 Jan 30];12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059.
- Brahmer JR, Abu-Sbeih H, Ascierto PA, et al. Society for immunotherapy of cancer (SITC) clinical practice guideline on immune checkpoint inhibitor-related adverse events. J Immunother Cancer [Internet]. 2021 [cited 2023 May 16];9(6):e002435. doi: 10.1136/jitc-2021-002435
- Shields MD, Marin-Acevedo JA, Pellini B. Immunotherapy for advanced non–small cell lung cancer: a decade of progress. Am Soc Clin Oncol Educ Book [Internet]. 2021 [cited 2023 Nov 9];41(41):e105–e127. Available from: http://www.ncbi.nlm.nih.gov/pubmed/33979196. doi: 10.1200/EDBK_321483
- Thapa B, Roopkumar J, Kim AS, et al. Incidence and clinical pattern of immune related adverse effects (irAE) due to immune checkpoint inhibitors (ICI). 2019;37:e14151–e14151.
- Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Long-term outcomes with nivolumab plus ipilimumab or nivolumab alone versus ipilimumab in patients with advanced melanoma. J Clin Oncol. 2022;40(2):127–137. doi: 10.1200/JCO.21.02229
- Arnaud-Coffin P, Maillet D, Gan HK, et al. A systematic review of adverse events in randomized trials assessing immune checkpoint inhibitors. Int J Cancer [Internet]. 2019 [cited 2023 May 16];145(3):639–648. doi: 10.1002/ijc.32132
- Horn LA, Fousek K, Palena C. Tumor plasticity and resistance to immunotherapy. Trends Cancer [Internet]. 2020 [cited 2023 Nov 2];6(5):432. Available from: /pmc/articles/PMC7192950/. doi: 10.1016/j.trecan.2020.02.001
- Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017
- Sankar K, Ye JC, Li Z, et al. The role of biomarkers in personalized immunotherapy. Biomark Res [Internet]. 2022 [cited 2024 Feb 11];10(1):1–13. doi: 10.1186/s40364-022-00378-0
- Meijer TG, Naipal KA, Jager A, et al. Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Sci OA [Internet]. 2017. cited 2022 Jan 22;3(2):FSO190. Available from: /pmc/articles/PMC5481868/. doi: 10.4155/fsoa-2017-0003
- Mason J, D Ö. Key aspects for conception and construction of co-culture models of tumor-stroma interactions. Front Bioeng Biotechnol. 2023;11:1150764. doi: 10.3389/fbioe.2023.1150764
- Hickman JA, Graeser R, de Hoogt R, et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J [Internet]. 2014 [cited 2024 Feb 11];9(9):1115–1128. doi: 10.1002/biot.201300492
- Du Y, Li X, Niu Q, et al. Development of a miniaturized 3D organoid culture platform for ultra-high-throughput screening. J Mol Cell Biol [Internet] 2020 [cited 2024 Feb 11];12:630. Available from: /pmc/articles/PMC7751183/. doi: 10.1093/jmcb/mjaa036.
- Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol [Internet]. 2003 [cited 2024 Feb 8];15(6):753. Available from: /pmc/articles/PMC2933200/. doi: 10.1016/j.ceb.2003.10.016
- Meyers J, Craig J, Odde DJJ. Potential for control of signaling pathways via cell size and shape. Curr Biol [Internet]. 2006 [cited 2024 Feb 8]. 16(17):1685–1693. doi: 10.1016/j.cub.2006.07.056.
- Mseka T, Bamburg JR, Cramer LP. Adf/cofilin family proteins control formation of oriented actin-filament bundles in the cell body to trigger fibroblast polarization. J Cell Sci [Internet]. 2007 [cited 2024 Feb 8]. 120(24):4332–4344. doi: 10.1242/jcs.017640.
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer [Internet]. 2005 [cited 2024 Feb 8]. 5(9):675–688. doi: 10.1038/nrc1695.
- Mahmud G, Campbell CJ, Bishop KJM, et al. Directing cell motions on micropatterned ratchets. Nat Phys [Internet]. 2009. cited 2024 Feb 8;5(8):606–612. Available from: https://www.nature.com/articles/nphys1306. doi: 10.1038/nphys1306
- Melissaridou S, Wiechec E, Magan M, et al. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int [Internet]. 2019 [cited 2024 Feb 8];19(1):1–10. doi: 10.1186/s12935-019-0733-1
- Zschenker O, Streichert T, Hehlgans S, et al. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS One [Internet]. 2012 [cited 2024 Feb 8];7(4):e34279. doi: 10.1371/journal.pone.0034279
- Krysko DV, Demuynck R, Efimova I, et al. In vitro veritas: from 2D cultures to organ-on-a-chip models to study immunogenic cell death in the tumor microenvironment. Cells [Internet]. 2022. cited 2024 Feb 11;11(22):3705. Available from: https://www.mdpi.com/2073-4409/11/22/3705/htm. doi: 10.3390/cells11223705
- Stock K, Estrada MF, Vidic S, et al. Capturing tumor complexity in vitro: comparative analysis of 2D and 3D tumor models for drug discovery. Sci Rep [Internet]. 2016 [cited 2024 Feb 11];6(1). Available from: /pmc/articles/PMC4929472/. doi: 10.1038/srep28951
- Mo X, Tang C, Niu Q, et al. HTiP: high-throughput immunomodulator phenotypic screening platform to reveal IAP antagonists as anti-cancer immune enhancers. Cell Chem Biol. 2019;26(3):331–339.e3. doi: 10.1016/j.chembiol.2018.11.011
- Mackenzie NJ, Nicholls C, Templeton AR, et al. Modelling the tumor immune microenvironment for precision immunotherapy. Clin Transl Immunology [Internet]. 2022 [cited 2024 Feb 11];11(6):e1400. doi: 10.1002/cti2.1400
- Olivo Pimentel V, Yaromina A, Marcus D, et al. A novel co-culture assay to assess anti-tumor CD8+ T cell cytotoxicity via luminescence and multicolor flow cytometry. J Immunol Methods. 2020;487:112899. doi: 10.1016/j.jim.2020.112899
- Rodrigues J, Heinrich MA, Teixeira LM, et al. 3D in vitro model (R)evolution: unveiling tumor–stroma interactions. Trends Cancer. 2021;7(3):249–264. doi: 10.1016/j.trecan.2020.10.009
- Campuzano S, Pelling AE. Scaffolds for 3D cell culture and cellular agriculture applications derived from non-animal sources. Front Sustain Food Syst. 2019;3:38. doi: 10.3389/fsufs.2019.00038
- Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17(1):1–15. doi: 10.1016/j.neo.2014.12.004
- Mäkelä R, Arjonen A, Härmä V, et al. Ex vivo modelling of drug efficacy in a rare metastatic urachal carcinoma. BMC Cancer [Internet] 2020 [cited 2021 Oct 3];20:1–10. Available from: https://bmccancer.biomedcentral.com/articles/10.1186/s12885-020-07092-w. doi: 10.1186/s12885-020-07092-w.
- Mönch D, Koch J, Maaß A, et al. A human ex vivo coculture model to investigate peritoneal metastasis and innovative treatment options. Pleura Peritoneum. 2021;6(3):121–129. doi: 10.1515/pp-2021-0128
- Tan SQ, Lee Y, Lee FYX, et al. Ex vivo co-culture models for immunotherapy with patient-derived tumor infiltrating lymphocytes, peripheral blood mononuclear cells and autologous patient colorectal cancer (CRC) cell lines. J Clin Oncol. 2018;36(15_suppl):e15531–e15531. doi: 10.1200/JCO.2018.36.15_suppl.e15531
- Gong X, Lin C, Cheng J, et al. Generation of multicellular tumor spheroids with microwell-based agarose scaffolds for drug testing. PLoS One [Internet]. 2015 [cited 2023 Jun 18];10(6):e0130348. doi: 10.1371/journal.pone.0130348
- Uchida N, Buck DW, He D, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci [Internet]. 2000 [cited 2023 Jun 18];97:p. 14720–14725. Available from: https://www.pnas.org/doi/abs/10.1073/pnas.97.26.14720.
- Kondo J, Endo H, Okuyama H, et al. Retaining cell–cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci U S A [Internet]. 2011. cited 2023 Jun 18;108(15):6235–6240. Available from: https://www.pnas.org/doi/abs/10.1073/pnas.1015938108. doi: 10.1073/pnas.1015938108
- Burgués JP, Gómez L, Pontones JL, et al. A chemosensitivity test for superficial bladder cancer based on three-dimensional culture of tumour spheroids. Eur Urol. 2007;51(4):962–970. doi: 10.1016/j.eururo.2006.10.034
- Powley IR, Patel M, Miles G, et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br J Cancer [Internet]. 2020. cited 2023 Jun 18;122(6):735–744. Available from: https://www.nature.com/articles/s41416-019-0672-6. doi: 10.1038/s41416-019-0672-6
- Datta P, Dey M, Ataie Z, et al. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis Oncol. 2020;4(1):4. doi: 10.1038/s41698-020-0121-2
- Ruppen J, Wildhaber FD, Strub C, et al. Towards personalized medicine: chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip [Internet]. 2015 [cited 2023 Oct 11];15(14):3076–3085. doi: 10.1039/C5LC00454C
- Herter S, Morra L, Schlenker R, et al. A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol Immunother [Internet]. 2017 [cited 2023 Nov 28];66(1):129–140. doi: 10.1007/s00262-016-1927-1
- Tong JG, Valdes YR, Barrett JW, et al. Evidence for differential viral oncolytic efficacy in an in vitro model of epithelial ovarian cancer metastasis. Mol Ther Oncolytics. 2015;2:15013. doi: 10.1038/mto.2015.13
- Mukherjee M, Chepizhko O, Chiara Lionetti M, et al. Infiltration of tumor spheroids by activated immune cells. Phys Biol [Internet]. 2023. cited 2023 Nov 28;20(5):056001. Available from: https://iopscience.iop.org/article/10.1088/1478-3975/ace0ee. doi: 10.1088/1478-3975/ace0ee
- He W, Kuang Y, Xing X, et al. Proteomic comparison of 3D and 2D glioma models reveals increased HLA-E expression in 3D models is associated with resistance to NK cell-mediated cytotoxicity. J Proteome Res [Internet]. 2014 [cited 2023 Nov 28];13(5):2272–2281. doi: 10.1021/pr500064m
- Courau T, Bonnereau J, Chicoteau J, et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J Immunother Cancer [Internet]. 2019 [cited 2023 Jul 28];7(1):1–14. doi: 10.1186/s40425-019-0553-9
- Ou L, Liu S, Wang H, et al. Patient-derived melanoma organoid models facilitate the assessment of immunotherapies. EBioMedicine [Internet]. 2023 [cited 2023 Nov 3];92:104614. Available from: http://www.thelancet.com/article/S2352396423001792/fulltext doi: 10.1016/j.ebiom.2023.104614.
- Wallstabe L, Göttlich C, Nelke LC, et al. ROR1-CAR T cells are effective against lung and breast cancer in advanced microphysiologic 3D tumor models. JCI Insight [Internet]. 2019 [cited 2023 Nov 3];4(18). doi: 10.1172/jci.insight.126345
- Neal JT, Li X, Zhu J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175(7):1972–1988.e16. doi: 10.1016/j.cell.2018.11.021.
- Mu H-Y, Lin C-M, Chu L-A, et al. Ex vivo evaluation of combination immunotherapy using tumor-microenvironment-on-chip. Adv Healthc Mater [Internet]. 2023 cited 2023 Nov 3;2302268. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/adhm.202302268
- Alexander Heinrich M, Bansal R, Lammers T, et al. 3D-Bioprinted mini-brain: a glioblastoma model to study cellular interactions and therapeutics. Adv Mater [Internet]. 2019 [cited 2024 Feb 12];31(14):1806590. doi: 10.1002/adma.201806590
- Maritan SM, Lian EY, Mulligan LM. An efficient and flexible cell aggregation method for 3d spheroid production. J Visualized Exp [Internet]. 2017 cited 2023 Nov 2;2017(121). doi: 10.3791/55544.
- Han SJ, Kwon S, Kim KS. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int [Internet]. 2021 [cited 2023 Jun 19];21(1):1–19. Available from: https://cancerci.biomedcentral.com/articles/10.1186/s12935-021-01853-8. doi: 10.1186/s12935-021-01853-8
- Barbosa MAG, Xavier CPR, Pereira RF, et al. 3D cell culture models as recapitulators of the tumor microenvironment for the screening of anti-cancer drugs. Cancers (Basel) [Internet]. 2021 [cited 2023 Jun 19];14(1):190. Available from: http://pmc/articles/PMC8749977/
- Weiswald LB, Richon S, Validire P, et al. Newly characterised ex vivo colospheres as a three-dimensional colon cancer cell model of tumour aggressiveness. Br J Cancer [Internet]. 2009. cited 2024 Feb 12;101(3):473. Available from: /pmc/articles/PMC2720229/. doi: 10.1038/sj.bjc.6605173
- Voabil P, de Bruijn M, Roelofsen LM, et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med [Internet]. 2021. cited 2024 Feb 12;27(7):1250–1261. Available from: https://www.nature.com/articles/s41591-021-01398-3. doi: 10.1038/s41591-021-01398-3
- Mu P, Zhou S, Lv T, et al. Newly developed 3D in vitro models to study tumor–immune interaction. J Exp Clin Cancer Res [Internet]. 2023 [cited 2024 Feb 12];42(1):1–16. doi: 10.1186/s13046-023-02653-w
- Demers I, Donkers J, Kremer B, et al. Ex vivo culture models to indicate therapy response in head and neck squamous cell carcinoma. Cells [Internet]. 2020. cited 2022 Jan 31;9(11):2527. Available from: /pmc/articles/PMC7700693/. doi: 10.3390/cells9112527
- Subia B, Dahiya UR, Mishra S, et al. Breast tumor-on-chip models: from disease modeling to personalized drug screening. J Controlled Release. 2021;331:103–120. doi: 10.1016/j.jconrel.2020.12.057
- Phung YT, Barbone D, Broaddus VC, et al. Rapid generation of in vitro multicellular spheroids for the study of monoclonal antibody therapy. J Cancer [Internet]. 2011 [cited 2023 May 16];2:507–514. doi: 10.7150/jca.2.507
- Huang YL, Ma Y, Wu C, et al. Tumor spheroids under perfusion within a 3D microfluidic platform reveal critical roles of cell-cell adhesion in tumor invasion. Sci Rep [Internet]. 2020. cited 2023 May 16;10(1):1–11. Available from: https://www.nature.com/articles/s41598-020-66528-2. doi: 10.1038/s41598-020-66528-2
- Kapałczyńska M, Kolenda T, Przybyła W, et al. 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Arch Med Sci [Internet]. 2018 [cited 2024 Feb 12];14: 910. Available from: http://pmc/articles/PMC6040128/.
- Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2013;18(5–6):240–249. doi: 10.1016/j.drudis.2012.10.003
- Benien P, Swami A. 3D tumor models: history, advances and future perspectives. Future Oncol [Internet]. 2014 [cited 2023 Jun 17];10(7):1311–1327. Available from: https://www.futuremedicine.com/doi/10.2217/fon.13.274
- Marques IA, Fernandes C, Tavares NT, et al. Magnetic-based human tissue 3D cell culture: a systematic review. IJMS [Internet]. 2022. cited 2024 Feb 13;23(20):12681. Available from: https://www.mdpi.com/1422-0067/23/20/12681/htm. doi: 10.3390/ijms232012681
- Ferreira JN, Hasan R, Urkasemsin G, et al. A magnetic three-dimensional levitated primary cell culture system for the development of secretory salivary gland-like organoids. J Tissue Eng Regen Med [Internet]. 2019 [cited 2024 Feb 13];13(3):495–508. doi: 10.1002/term.2809
- Chan WS, Mo X, Ip PPC, et al. Patient-derived organoid culture in epithelial ovarian cancers—techniques, applications, and future perspectives. Cancer Med [Internet]. 2023 [cited 2024 Feb 13];12(19):19714–19731. doi: 10.1002/cam4.6521
- Wang Y, Shi W, Kuss M, et al. 3D bioprinting of breast cancer models for drug resistance study. ACS Biomater Sci Eng [Internet]. 2018. cited 2024 Feb 13;4(12):4401–4411. Available from: https://pubs.acs.org/doi/full/10.1021/acsbiomaterials.8b01277. doi: 10.1021/acsbiomaterials.8b01277
- Yadav B, Pemovska T, Szwajda A, et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep [Internet]. 2014. cited 2023 Nov 28;4(1):1–10. Available from: https://www.nature.com/articles/srep05193. doi: 10.1038/srep05193
- Thoma CR, Stroebel S, Rösch N, et al. A high-throughput–compatible 3D microtissue co-culture system for phenotypic RNAi screening applications. SLAS Discovery [Internet]. 2013. cited 2023 Jun 19;18(10):1330–1337. Available from: https://pubmed.ncbi.nlm.nih.gov/24080258/. doi: 10.1177/1087057113499071
- Zanoni M, Piccinini F, Arienti C, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci Rep [Internet]. 2016 [cited 2023 Jun 19];6(1). Available from: /pmc/articles/PMC4707510/. doi: 10.1038/srep19103
- Särchen V, Shanmugalingam S, Kehr S, et al. Pediatric multicellular tumor spheroid models illustrate a therapeutic potential by combining BH3 mimetics with natural killer (NK) cell-based immunotherapy. Cell Death Discov [Internet]. 2022. cited 2023 Jun 19;8(1):1–10. Available from: https://www.nature.com/articles/s41420-021-00812-6. doi: 10.1038/s41420-021-00812-6
- Wan C, Keany MP, Dong H, et al. Enhanced efficacy of simultaneous PD-1 and PD-L1 immune checkpoint blockade in high-grade serous ovarian cancer. Cancer Res [Internet]. 2021. cited 2023 Jun 19;81(1):158. Available from: /pmc/articles/PMC7878408/. doi: 10.1158/0008-5472.CAN-20-1674
- Meng Q, Xie S, Gray GK, et al. Empirical identification and validation of tumor-targeting T cell receptors from circulation using autologous pancreatic tumor organoids. J Immunother Cancer. [Internet] 2021 [cited 2023 Jun 19];9(11):e003213. doi: 10.1136/jitc-2021-003213
- Zhou S, Zhu M, Meng F, et al. Evaluation of PD-1 blockade using a multicellular tumor spheroid model. Am J Transl Res [Internet]. 2019 [cited 2023 Nov 16];11: 7471. Available from: http://pmc/articles/PMC6943460/.
- Sirenko O, Mitlo T, Hesley J, et al. High-content assays for characterizing the viability and morphology of 3D cancer spheroid cultures. Assay Drug Dev Technol [Internet]. 2015. cited 2023 Nov 3;13(7):402. Available from: /pmc/articles/PMC4556086/. doi: 10.1089/adt.2015.655
- Kast V, Nadernezhad A, Pette D, et al. A tumor microenvironment model of pancreatic cancer to elucidate responses toward immunotherapy. Adv Healthc Mater [Internet] 2023. cited 2023 Jul 28;12(14):2201907. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/adhm.202201907. doi: 10.1002/adhm.202201907
- Booij TH, Price LS, Danen EHJ. 3D cell-based assays for drug screens: challenges in imaging, image analysis, and high-content analysis. SLAS DISCOVERY [Internet]. 2019 [cited 2023 Nov 3];24(6):615–627. doi: 10.1177/2472555219830087.
- Rolver MG, Elingaard-Larsen LO, Pedersen SF. Assessing cell viability and death in 3d spheroid cultures of cancer cells. J Visualized Exp. 2019;2019(148). doi: 10.3791/59714.
- Edwards SJ, Carannante V, Kuhnigk K, et al. High-resolution imaging of tumor spheroids and organoids enabled by expansion microscopy. Front Mol Biosci. 2020;7:567282. doi: 10.3389/fmolb.2020.00208
- Diosdi A, Hirling D, Kovacs M, et al. Cell lines and clearing approaches: a single-cell level 3D light-sheet fluorescence microscopy dataset of multicellular spheroids. Data Brief. 2021;36:107090. doi: 10.1016/j.dib.2021.107090
- Saraiva DP, Matias AT, Braga S, et al. Establishment of a 3D co-culture with MDA-MB-231 breast cancer cell line and patient-derived immune cells for application in the development of immunotherapies. Front Oncol. 2020;10:561826. doi: 10.3389/fonc.2020.01543
- Zhang Z, Wang G, Zhong K, et al. A drug screening to identify novel combinatorial strategies for boosting cancer immunotherapy efficacy. J Transl Med [Internet]. 2023 [cited 2023 Apr 5];21(1):1–20. doi: 10.1186/s12967-023-03875-4
- Ruppen J, Cortes-Dericks L, Marconi E, et al. A microfluidic platform for chemoresistive testing of multicellular pleural cancer spheroids. Lab Chip [Internet]. 2014 [cited 2023 Jun 19];14(6):1198–1205. doi: 10.1039/C3LC51093J
- Li C, Holman JB, Shi Z, et al. On-chip modeling of tumor evolution: advances, challenges and opportunities. Mater Today Bio. 2023;21:100724. doi: 10.1016/j.mtbio.2023.100724
- Li NT, Co IL, Landon-Brace N, et al. Tissue-engineered 3D cancer microenvironment for screening therapeutics. In: Kundu SC, Reis RL, editors. Biomate 3D Tumor Model. Elsevier; 2020. p. 453–479. doi: 10.1016/B978-0-12-818128-7.00019-8
- Meindl-Beinker NM, Betge J, Gutting T, et al. A multicenter open-label phase II trial to evaluate nivolumab and ipilimumab for 2nd line therapy in elderly patients with advanced esophageal squamous cell cancer (RAMONA). BMC Cancer [Internet]. 2019 [cited 2024 Feb 13];19(1). Available from: /pmc/articles/PMC6419339/. doi: 10.1186/s12885-019-5446-2
- Snijder B, Vladimer GI, Krall N, et al. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. 2017 [cited 2023 Nov 5]; Available from: http://www.thelancet.com/haematologyPublishedonline.
- Kropivsek K, Kachel P, Goetze S, et al. Ex vivo drug response heterogeneity reveals personalized therapeutic strategies for patients with multiple myeloma. Nat Cancer. 2023. cited 2023 Aug 2;4(5):734–753. Available from: https://www.nature.com/articles/s43018-023-00544-9. doi: 10.1038/s43018-023-00544-9
- Smiraglia DJ, Rush LJ, Frühwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Human Molecular Genetics [Internet]. 2001. cited 2023 Nov 5;10(13):1413–1419. Available from: https://pubmed.ncbi.nlm.nih.gov/11440994/. doi: 10.1093/hmg/10.13.1413
- Pan C, Kumar C, Bohl S, et al. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol & Cell Proteom [Internet]. 2009. cited 2023 Nov 5;8(3):443. Available from: /pmc/articles/PMC2649808/. doi: 10.1074/mcp.M800258-MCP200
- Li YR, Yu Y, Kramer A, et al. An ex vivo 3D tumor microenvironment-mimicry culture to study TAM modulation of cancer immunotherapy. Cells [Internet]. 2022. cited 2023 Nov 5;11(9):1583. Available from: /pmc/articles/PMC9101510/. doi: 10.3390/cells11091583
- Lee SH, Hu W, Matulay JT, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173(2):515–528.e17. doi: 10.1016/j.cell.2018.03.017
- Grassi L, Alfonsi R, Francescangeli F, et al. Organoids as a new model for improving regenerative medicine and cancer personalized therapy in renal diseases. Cell Death Dis [Internet]. 2019. cited 2023 Nov 6;10(3):1–15. Available from: https://www.nature.com/articles/s41419-019-1453-0. doi: 10.1038/s41419-019-1453-0
- Martini G, Belli V, Napolitano S, et al. Establishment of patient-derived tumor organoids to functionally inform treatment decisions in metastatic colorectal cancer. ESMO Open. 2023;8(3):101198. doi: 10.1016/j.esmoop.2023.101198
- Subtil B, Iyer KK, Poel D, et al. Dendritic cell phenotype and function in a 3D co-culture model of patient-derived metastatic colorectal cancer organoids. Front Immunol. 2023;14:1105244. doi: 10.3389/fimmu.2023.1105244
- Longati P, Jia X, Eimer J, et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer [Internet]. 2013 [cited 2023 Jun 18];13(1):1–13. doi: 10.1186/1471-2407-13-95
- Ryu NE, Lee SH, Park H. Spheroid culture system methods and applications for mesenchymal stem cells. Cells [Internet]. 2019 [cited 2023 Jun 18];8(12):1620. Available from: https://www.mdpi.com/2073-4409/8/12/1620/htm. doi: 10.3390/cells8121620
- Białkowska K, Komorowski P, Bryszewska M, et al. Spheroids as a type of three-dimensional cell cultures—examples of methods of preparation and the most important application. Int J Mol Sci [Internet]. 2020. cited 2023 Jun 18;21(17):6225. Available from. https://www.mdpi.com/1422-0067/21/17/6225/htm. doi: 10.3390/ijms21176225
- Wang T, Wang L, Wang G, et al. Leveraging and manufacturing in vitro multicellular spheroid-based tumor cell model as a preclinical tool for translating dysregulated tumor metabolism into clinical targets and biomarkers. Bioresour Bioprocess [Internet]. 2020. cited 2023 Jun 17;7(1):1–34. Available from: https://bioresourcesbioprocessing.springeropen.com/articles/10.1186/s40643-020-00325-7. doi: 10.1186/s40643-020-00325-7
- Gunti S, Hoke ATK, Vu KP, et al. Organoid and spheroid tumor models: techniques and applications. Cancers (Basel) [Internet]. 2021. cited 2023 Jun 17;13(4):1–18. Available from: /pmc/articles/PMC7922036/. doi: 10.3390/cancers13040874
- Unnikrishnan K, Thomas LV, Ram Kumar RM. Advancement of scaffold-based 3D cellular models in cancer tissue engineering: an update. Front Oncol [Internet]. 2021 [cited 2023 Jun 17];11.Available from: /pmc/articles/PMC8573168/. doi: 10.3389/fonc.2021.733652

