ABSTRACT
Colorectal cancer (CRC) is a prevalent tumour with high morbidity rates worldwide, and its incidence among younger populations is rising. Early diagnosis of CRC can help control the associated mortality. Fungi are common microorganisms in nature. Recent studies have shown that fungi may have a similar association with tumours as bacteria do. As an increasing number of tumour-associated fungi are discovered, this provides new ideas for the diagnosis and prognosis of tumours. The relationship between fungi and colorectal tumours has also been recently identified by scientists. Therefore, this paper describes the limitations and prospects of the application of fungi in diagnosing CRC and predicting CRC prognosis.
1. Introduction
Colorectal cancer (CRC) is a heterogeneous disease characterised by genetic and epigenetic abnormalities (Ashktorab and Brim Citation2022; Joanito et al. Citation2022). It can occur in any part of the colon or rectum and spread to other tissues and organs through lymphatic circulation, blood circulation, and direct transmission (Galindo-Pumarino et al. Citation2021). Currently, CRC is the second leading cause of cancer-related mortality worldwide (Sung et al. Citation2021). Furthermore, the burden of CRC in many countries, including China, which has a large population (Collaborators GBDCC Citation2022; Qu et al. Citation2022), has increased each year and is influenced by lifestyle (Abu-Ghazaleh et al. Citation2021; Qin et al. Citation2022) and other factors. There is evidence that the incidence rate and mortality of early-onset colorectal neoplasms (i.e. colorectal neoplasms diagnosed in patients under 50 years old) are increasing worldwide (O’Sullivan et al. Citation2022; Patel et al. Citation2022). Fortunately, the development of CRC is not rapid (Yelorda et al. Citation2021). A population-based global multicenter study suggests that when the proportion of early-stage CRC among cases identified by screening is large, patient prognosis is better (Cardoso et al. Citation2022). Therefore, more attention should be given to the early screening of colorectal tumours.
Endoscopy (including colonoscopy and sigmoidoscopy), stool tests [including faecal occult blood test (FOBT), faecal immunochemical test (FIT), and FIT-DNA tests], and plasma SEPT9 gene methylation are widely used in clinical practice as prevention strategies and screening programmes for CRC, as supported by scholars (Kanth and Inadomi Citation2021; Li et al. Citation2021). However, it is worth noting that despite the high false positive rate of non-invasive tests currently used in clinical practice, patient compliance with undergoing non-invasive tests is better than that for invasive tests in the population (Ballester et al. Citation2022). A study on the correlation between risk factors and intervention strategies for CRC demonstrated that non-invasive testing is more cost-efficient for the general public and more accessible in primary care settings, reducing the risk of death due to CRC (Carethers and Doubeni Citation2020). Recently, a joint European multi-association guideline recommended the use of FIT to prioritise the treatment of patients with clinical characteristics of CRC in primary care facilities (Monahan et al. Citation2022). However, retrospective studies have also confirmed the role of endoscopy. A nationwide cohort study in South Korea suggests that colonoscopy can lower the risk of CRC more than FIT (Sung et al. Citation2022). These studies not only prompt us to improve visual research strategies but also urge us to identify other non-invasive inspection strategies with higher sensitivity and specificity (Chung et al. Citation2022).
Fungi and bacteria are widely distributed in nature and are also commonly present in the human oral cavity (Mukherjee and Leys Citation2021; Cheung et al. Citation2022), stomach (Liu et al. Citation2022), and intestines (Fassarella et al. Citation2021; Nel Van Zyl et al. Citation2022). The intestinal flora, which is an important component of the human intestinal microbiota, has been shown to have a profound impact on CRC, as evidenced by numerous studies (Kvaerner et al. Citation2021; Rebersek Citation2021; Clay et al. Citation2022; Shi et al. Citation2023). In recent years, an increasing number of tumour-associated fungi have been discovered, which provides new ideas for the diagnosis and prognosis of tumours.
The purpose of this review is to discuss recent advances in the diagnosis of CRC by fungi.
2. Fungi and tumours
2.1. Is fungi related to the occurrence and development of gastrointestinal tumors?
Fungi, bacteria, and viruses are ubiquitous microorganisms. Investigating the relationship between microorganisms and gastrointestinal tumours has been the subject of numerous studies. Many studies have demonstrated that bacterial infections, such as Helicobacter pylori infections, are linked to the development of gastrointestinal tumours such as gastric cancer (Smyth et al. Citation2020), while other studies have found that Fusobacterium nucleatum infection is associated with CRC (Clay et al. Citation2022; Wang and Fang Citation2023). Some researchers have linked Epstein-Barr virus (EBV) to stomach cancer (Zheng et al. Citation2022; Noh et al. Citation2023). Moreover, several viruses have been described in the context of CRC, such as EBV, human tumour virus (HPV), John Cunningham virus (JCV), and bacteriophages (Marongiu and Allgayer Citation2022; Sarantis et al. Citation2022).
In the field of fungi research, earlier investigators speculated that the fungal flora may be associated with or even promote pancreatic cancer (Dambuza and Brown Citation2019; Hindson Citation2019). Building on this, additional researchers have further found that fungi can promote IL-33 secretion and type 2 immune responses that lead to the development and progression of pancreatic cancer (Alam et al. Citation2022). A recent analysis of the cancerous and adjacent noncancerous tissues of 45 patients with gastric cancer in Shenyang, China, revealed that Candida species, especially Candida albicans (C. albicans), are enriched in gastric cancer tissue (Zhong et al. Citation2021). However, some scholars have summarised previous studies on the correlation between the gastric microbiota and gastric cancer and believe that it is still uncertain whether there is a correlation between the diversity of the gastric microbiota and the transformation of healthy gastric mucosa into gastric cancer (Stewart et al. Citation2020).
A population study has shown that patients with confirmed CRC who receive the antifungal agent terbinafine have a lower risk of death (hazard ratio = 0.50) and metastasis (hazard ratio = 0.44) than patients who do not take terbinafine (Hu et al. Citation2022). The mouse experiments conducted during the same project also demonstrated that terbinafine can inhibit CRC by influencing fungi (Hu et al. Citation2022). The researchers found significant changes in the proportion of species in the fungal community composition of the mice treated with terbinafine (Hu et al. Citation2022). Another mouse trial also demonstrated that terbinafine can be used synergistically with other anticancer drugs to treat CRC (Li et al. Citation2022). Researchers have proposed that disorders of the intestinal flora, particularly the fungal flora, promote CRC by affecting intestinal barrier function (Li et al. Citation2022). In addition, the two animal experiments described above indicated that terbinafine can directly inhibit the proliferation of CRC cells by targeting squalene epoxidase, a rate-limiting enzyme in cholesterol biosynthesis (Hu et al. Citation2022; Li et al. Citation2022). Mouse experiments further indicated the relationship between fungi and gastrointestinal tumours. Other researchers have found that treating Dectin-3 deficient mice with fluconazole prevented colitis‐associated colon cancer (CAC) progression. They also observed an increase in C. albicans abundance in Dectin-3-deficient mice, which subsequently promoted CAC development (Zhu et al. Citation2021). These findings provide a foundation for further exploring the relationship between C. albicans and CRC.
2.2. Candida and gastrointestinal tumours
Candida is the most prevalent and abundant fungus in the gastrointestinal tract and on other mucosal surfaces in humans and several other animals (Perez Citation2021). C. albicans is the main cause of candidiasis worldwide (Bilal et al. Citation2022). Other common disease-related Candida species, such as Candida parapsilosis (Branco et al. Citation2023), Candida tropicalis (Xu Citation2021), and Candida auris (Bing et al. Citation2022; Du et al. Citation2022), are also concerning. Recent research has revealed that digestive tract tumours (including gastric and intestinal cancers) contain high levels of C. albicans, which differs from other nondigestive tract tumours (Dohlman et al. Citation2022). In CRC, tumour metastasis and poor prognosis are more likely to occur as the C. albicans content increases. We need to further explore the relationship between Candida and CRC in greater detail.
Recently, some studies have shown that inflammatory genes are novel predictive markers of CRC, highlighting the role of inflammation in CRC (Schmitt and Greten Citation2021). Researchers have found that in gastrointestinal tumours with a high concentration of Candida, inflammatory responses can promote colonisation by Candida, while Candida itself can maintain an inflammatory environment, which is associated with the activation of inflammatory signalling pathways mediated by the proinflammatory factor IL-1 and neutropenia (Dohlman et al. Citation2022). In another study, it was discovered that C. albicans could activate glycolysis and IL‐7 production in macrophages, suggesting that the fungus is associated with the inflammatory response of macrophages (Zhu et al. Citation2021). Some experiments in mice have shown that strains of the fungi that produce candidalysin induce an increase in the number of interleukin-17A-producing T helper cell (Th 17 cell) cells and other immune cells involved in inflammation, such as neutrophils (Li et al. Citation2022). Other studies have reported that C. tropicalis induces CRC by activating NLRP3 inflammatory bodies through glycogen metabolism-dependent glycolysis and JAK-STAT1 signalling pathways (Xu et al. Citation2022). Additionally, there is growing evidence that patients undergoing cholecystectomy may be at higher risk of postoperative gastrointestinal complications (such as CRC), which may be related to Candida glabrata (Xu et al. Citation2022). It can be suspected that Candida in the cells of CRC tumours participates in the occurrence and development of CRC by causing inflammation.
Several studies have confirmed the potential link between Candida infection and oral squamous cell carcinoma (OSCC), although the specific mechanism has not yet been fully defined (Stasiewicz and Karpinski Citation2022). After a comprehensive analysis of the spatial distribution of microorganisms in tissue samples of OSCC and CRC (Galeano Nino et al. Citation2022), it was speculated that the distribution of microbial populations within tumours was not random. In contrast, the microbial community exhibits a highly organised structure with immune and epithelial functions that promote cancer progression.
There is not much evidence to prove that Candida directly causes gastrointestinal tumours, but it appears to work through the host’s immune system and intestinal epithelial function.
2.3. Fungi and inflammatory bowel disease (IBD)
IBD is a high-risk factor for CRC (Shah and Itzkowitz Citation2022). Recent studies have proposed the role of fungi in IBD (Iliev and Cadwell Citation2021). Several studies using ITS2 sequencing to analyse the fungal composition of the faecal microbiota have identified Candida species as the main genus responsible for IBD (Wang et al. Citation2021, Citation2023). In addition, some studies have revealed that intestinal fungi promote the occurrence of IBD (Krawczyk et al. Citation2023), which may be related to the Dectin-1- SYK-CARD9/NF-κB signalling pathway (Zajta et al. Citation2021; Yu et al. Citation2023). More interestingly, recent observations indicate the involvement of the Dectin-1-PGE2-IL-22BP axis in regulating intestinal tumorigenesis. Dectin-1, encoded by the Clec7a gene, is known to play important roles in host defence against fungi and immune homoeostasis in the intestine and will likely be a therapeutic target in the future (Tang et al. Citation2023). Therefore, the assessment of fungal-related biomarkers may also help identify risk factors for CRC in combination with other non-invasive examination methods (Pratt et al. Citation2022).
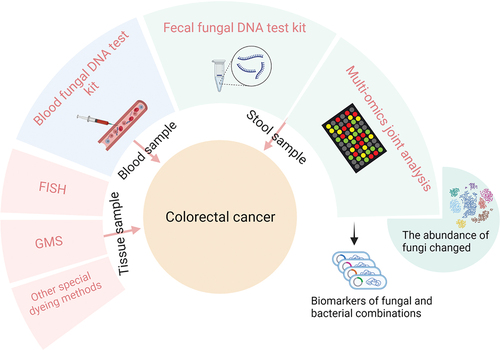
3. Fungal signatures of CRC identified using different samples
3.1. Fungal signatures in tumours
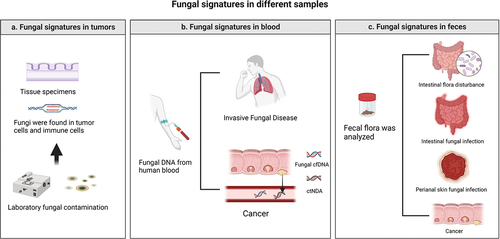
In previous work, Nejman and other scientists discovered the presence of metabolically active, immunologically active, intracellular, and cancer type-specific bacteria in tumour tissue. Most of these bacteria impact cancer treatment and are therefore included in the new cancer signature (Greathouse et al. Citation2020; Nejman et al. Citation2020). Due to the low abundance of fungi in tumours and the difficulty of sample purification, there is a lack of comprehensive reporting on the fungal community in various tumours. More recently, Narunsky-Haziza et al. (Citation2022) drew on the work of Nejman et al. to compare and analyse fungal communities with matched bacterial groups and immune groups, further suggesting the presence of not only bacteria but also fungi in tumours and highlighting fungi as potential biomarkers for tumour diagnosis. The study revealed that fungi and bacteria have similarities, with most hiding within cancer cells or immune cells inside tumours. Therefore, fungi and their metabolism may affect not only cancer cells but also immune cells and their activity, which may provide us with new ideas for targeted treatment. The discovery of specific fungi in tumours can therefore complement biopsy tests, not only for disease diagnosis but also to determine the origin of tumours, identify tumour subtypes and guide subsequent treatment. In addition, it is important to be aware of interference from common laboratory fungal contamination in the results of oncological pathology (the diseases that may require differential diagnosis are shown in ).
Figure 1. Diseases that require differential diagnosis. (a) Laboratory fungal infections should be considered during biopsy of tumour tissue. (b) Tumor patients are often immunosuppressed after chemotherapy and radiotherapy and are prone to invasive mycosis. Therefore, fungi and fungal antibodies can also be found in the blood. (c) If a change in faecal fungal flora abundance is found, in addition to considering CRC, intestinal flora disorders, fungal intestinal infections, and perianal skin infections should also be considered.
3.2. Fungi in blood
3.2.1. Other CRC markers in bodily fluids
Liquid biopsy analysis of circulating biomarkers, such as circulating tumour DNA (ctDNA) (Nassar et al. Citation2021) and exosomes (Elmallah et al. Citation2022; Zheng et al. Citation2022), for CRC have also received increasing attention (Mauri et al. Citation2022; Zhou et al. Citation2022). These biomarkers have the advantage of better-predicting disease progression in addition to being useful for screening. Previous studies have proposed that the microbial cell-free DNA (mcfDNA) can be used to distinguish individuals with some tumours from healthy populations, and this biomarker may enable CRC screening and early diagnosis (Ajami and Wargo Citation2020; Poore et al. Citation2020; Xiao et al. Citation2021). The SEPT9 gene methylation test has also been approved by the United States Food and Drug Administration (US FDA) for screening for CRC in the US (Kamel et al. Citation2022).
3.2.2. Fungal signatures in blood
In a pan-cancer analysis, fungi from 17,401 samples from patients with 35 types of cancer were investigated, and the first pan-cancer mycology atlas was generated (Narunsky-Haziza et al. Citation2022). By testing blood samples from cancer patients, researchers have found that specific fungal and bacterial DNA exists in human blood. These detected fungal DNA components are only related to specific types of tumours and have no clear relationship with the tumour stage. This means that we may be able to detect CRC in individuals who do not exhibit clinical symptoms or even adenoma before detecting the cancer stage. Aberrant ctDNA methylation and exosomal microRNA markers also have promising applications in the early detection of colorectal cancer, and mcfDNA can be combined with these liquid biopsy methods for colorectal cancer screening (Poore et al. Citation2020; Zhao et al. Citation2020). Another advantage of assessing microbial DNA-based cancer detection is that there is a considerable diversity of microbial DNA throughout different parts of the body (Narunsky-Haziza et al. Citation2022). Previous work suggests that it may be possible to accurately detect the presence and type of cancer at an early stage by using fungal signature information in the blood.
3.2.3. Fungi and invasive fungal disease (IFD)
Fungi are a ubiquitous class of organisms, and some can cause infections in humans. IFD, also known as invasive fungal infection (IFI), refers to disease-causing fungal invasion of human tissues and blood (Bassetti et al. Citation2021; Lass-Florl et al. Citation2021). The presence of new anticancer therapies has led to an increase in the number of immunosuppressed individuals, resulting in a higher diversity of fungal infections and an increase in the number of patients with IFD associated with cancer (Vehreschild et al. Citation2021; Rayens et al. Citation2022). Candida (McCarty et al. Citation2021) and Aspergillus (Foppiano Palacios and Spichler Moffarah Citation2021) are commonly identified causative pathogens in IFD.
However, the identities of pathogens that commonly cause IFD can overlap with specific fungal species found in CRC tissue. Consequently, there is a risk of false positive diagnosis when fungal biomarkers in the blood are used to differentiate and diagnose IFD and CRC patients. Therefore, to improve the accuracy of diagnosis, we may need to combine other biomarkers and auxiliary methods.
3.3. Fungal signatures in faeces
3.3.1. Fecal examination
(1) The FOBT uses chemical analyses to detect trace amounts of blood in faeces that are invisible to the naked eye. Its sensitivity in the detection of CRC and precancerous lesions is low and influenced by diet and drugs. In clinical practice, chemical, and immune methods are commonly used in combination (Kaur et al. Citation2023). (2) The FIT is used to detect human haemoglobin in faecal samples through specific antibodies (Randel et al. Citation2021). (3) Multitarget faecal DNA for detecting DNA mutations in faecal exfoliated cells can also be used in conjunction with FIT (Anderson et al. Citation2022; Xu et al. Citation2022).
3.3.2. Fungal signatures and CRC
Several teams from Shanghai recently collaborated on a project involving shotgun metagenomic sequencing of faecal samples from CRC patients in multiple geographical regions, identifying four kingdoms of microorganisms, including bacteria, fungi, archaea, and viruses (Liu et al. Citation2022). The random forest analysis they performed revealed that the model comprising only fungal markers had a higher performance in predicting CRC diagnosis than the model comprising only bacterial markers in two datasets. Notably, the combined analysis of bacterial and fungal markers showed greater accuracy than the analysis of only fungal or bacterial markers, providing solid evidence of the value of the non-invasive assessment of faecal CRC biomarkers. Simultaneously, another team of researchers analysed a faecal macrogenetic dataset obtained from 1,329 patients (454 CRC patients, 350 adenoma patients, and 525 healthy individuals) and found that the combined analysis of bacterial and fungal biomarkers more accurately distinguished between CRC patients and healthy individuals than the analysis of only bacterial biomarkers, increasing the area under the curve by 1.44% to 10.60%. Additionally, in vivo and in vitro experiments confirmed the association of Aspergillus with CRC occurrence (Lin et al. Citation2022). Previous scholars have also described multikingdom diagnostic models (Dickson Citation2019). These studies suggest that the combined analysis of bacterial and fungal biomarkers has considerable potential for the early screening of CRC. However, the current concept of using fungal biomarkers in stool is solely based on the understanding of the relationship between the intestinal microbiota and CRC and does not consider the relationship between tumour type-specific intracellular fungi and the occurrence of CRC.
Faecal microbiota transplantation (FMT), which is a method for treating diseases by transferring healthy faecal microbiota to the recipient, is one potential method for managing CRC. Recent studies have emphasised the role of viruses and fungi, such as bacteriophages and Candida, in addition to bacteria, in the efficacy of FMT (Shen et al. Citation2021; Lam et al. Citation2022). Furthermore, several studies suggest that modulating the intestinal microbiota through FMT can prevent and treat CRC (Chang et al. Citation2020; Huang et al. Citation2022). This again highlights the relationship between the intestinal microbiota and CRC.
3.3.3. Models combining bacterial and fungal features
New research urges us to reconsider the role of interactions between bacteria and fungi in CRC, as this concept has been repeatedly proposed (Lapiere and Richard Citation2022). Studies have demonstrated that interactions between bacteria and fungi can induce CRC pathogenesis by activating D-arginine and D-ornithine pathways and butanoate metabolism (Liu et al. Citation2022). Furthermore, it has been suggested that Lactobacillus rhamnosus is associated with the pathogenicity of the specific fungus C. albicans found in CRC tumour tissue (Alonso-Roman et al. Citation2022). In a longitudinal study of 178 premature infants, important interactions between bacteria and fungi were found in the gut beginning at birth (Rao et al. Citation2021). However, some studies suggest that the interaction between bacteria and fungi elicits limited effects, such as in the oral cavity (Cheung et al. Citation2022). These studies suggest that relationships between bacteria and fungi do not solely involve symbiosis, competition, or coordination and that the mechanisms and targets of action require further investigation. The current state of the research suggests that we need more clinical evidence (the evaluation of some faecal microbial markers is provided in ).
Table 1. Evaluation of some colorectal markers associated with microorganisms.
4. Techniques for detecting fungi
With advances in technology for assessing the microbiome, such as metagenomics, metabolomics, metaproteomics, and metatranscriptomics, a better understanding of intratumoral fungi has emerged (Guo et al. Citation2021; Lind and Pollard Citation2021; Dohlman et al. Citation2022; Ko et al. Citation2022; Kong and Machida Citation2022; Narunsky-Haziza et al. Citation2022; Caesar et al. Citation2023). In addition to identifying species, these techniques can provide insights into the metabolites of fungi. Analysing fungal metabolites can reduce the influence of dead microbes better than analysing fungal genes. However, metagenomic sequencing technology is more expensive than traditional amplicon sequencing technology. For fungi, we can use 18S rRNA gene amplicon sequencing and internal transcribed spacer (ITS) sequencing (Debeljak and Baltar Citation2023; Moreira et al. Citation2023).
Identifying fungi is generally difficult, but detecting fungi in tumours is also a challenge. There is no single way to detect all fungi. In addition, fungi in tumours are present in such small numbers that they are even more difficult to detect than bacteria in tumours, which makes identifying tumours through fungi a considerable challenge. For the detection of fungi, a microbial detection method with a low detection threshold is needed. Fluorescence in situ hybridisation (FISH) is a popular microbial detection technique (Liu et al. Citation2021; Sampaio et al. Citation2022). To detect intestinal mucosal microorganisms, CLAS-fish, BONCAT-FISH, HiPR-FISH, seq-FISH, and other techniques have been developed (Barbosa et al. Citation2023). However, we need to develop new probes for a wider range of fungi (Petriglieri et al. Citation2022). Narunsky-Haziza et al. (Citation2022) integrated four staining methods with varying sensitivity and specificity in the above experiments to improve the accuracy and efficiency of fungal detection, mainly based on Grocott’s methenamine silver (GMS) stain, cell wall antibodies, and autoantibodies, and FISH. Notably, the origin of the fungus in the tumour is also unclear. Scientists have explored the possible origin of bacteria in tumours from mucosal sites, normal adjacent tissues, and the circulatory system (Xie et al. Citation2022). Whether fungi come from places similar to the origins of bacteria still needs to be explored. Because the mechanisms through which the fungus circulates throughout the body and within specific organs and tissues remain unclear, technical limitations and a lack of understanding have hindered the specific detection and localisation of microorganisms within tumours (Strickland and Shi Citation2021).
The gold standard for the diagnosis of fungal infections remains the visualisation of fungal elements in samples from usually sterile sites (Borman et al. Citation2022). Although metagenomics has shown great promise in the study of both fungal infection and the fungi associated with tumours, microscopic examination and direct culture are still very important methods for obtaining evidence (Kamau and Yang Citation2023). At present, there is no real indicator with independent diagnostic value, so we tend to use a combination of multiple methods to improve the accuracy of fungal diagnosis. Therapeutic approaches such as FMT continue to inspire us to study the aspects of tumour-associated microorganisms in pure cultures, which remains important because the results of in vitro and in vivo experiments based on microbial cultures not only provide more reliable evidence but can also be used to support further treatment (Yu et al. Citation2023).
Moreover, because fungi are prevalent in nature, which makes sample contamination a serious problem, researchers must be very careful to filter out any potential contamination from the results (Narunsky-Haziza et al. Citation2022). In the study of fungi in tumours, contamination by host DNA and environmental microbial DNA is also an obstacle. We can consider establishing and using a contaminant-controlled analysis framework (Zozaya-Valdes et al. Citation2021). Dohlman et al. developed a decontamination algorithm that can remove contamination from The Cancer Genome Atlas (TCGA) data (Dohlman et al. Citation2021, Citation2022).
Overall, to date, there are no experimental methods that are both reliable and cost-effective for detecting fungi ().
5. Conclusions and outlook
Numerous studies have investigated the role of the tumour microbiota in tumour formation, development, and treatment, but comparatively limited attention has been given to studies on fungi in tumours. This review aims to address this gap and suggest the potential for fungi as a diagnostic and prognostic tool for CRC. The latest understanding of microorganisms within tumours is largely due to the development of techniques to detect microbial species and their metabolites. However, despite advancements in our understanding of the number of fungi in tumours and the relationship between tumours and fungi, more specific and sensitive techniques are necessary.
The use of fungi as biomarkers for CRC has considerable potential. However, several considerations must be addressed. First, for tumour patients with an immunosuppressive condition and concurrent fungal infections, such as fungemia or fungal enteritis, the diagnostic value of fungal biomarkers may decrease. Thus, further investigation is necessary to distinguish between the two conditions. Additionally, a more in-depth understanding of the relationship between fungi and CRC prognosis is essential for utilising fungi to assess CRC prognosis. This method offers the advantage of dynamically monitoring patients’ conditions, surpassing endoscopy and other currently used diagnostic methods. However, there is a lack of animal model experiments and clinical data to explain the relationship between fungi and CRC occurrence and development. Nonetheless, the strategy of diagnosing CRC through fungi is undoubtedly worthy of further consideration.
Author contributions
Xu-Huan Li and Ming-Ming Luo: Conceptualisation, Formal analysis, Visualisation, Writing-Original Draft, and Review & Editing. Zu-Xiu Wang: Visualisation, Review, and Editing. Qi Wang and Bin Xu: Conceptualisation, Funding acquisition, Supervision, Writing – Review & Editing.
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Funding
References
- Abu-Ghazaleh N, Chua WJ, Gopalan V. 2021. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J Gastroenterol Hepatol. 36(1):75–88. doi: 10.1111/jgh.15042.
- Ajami NJ, Wargo JA. 2020. AI finds microbial signatures in tumours and blood across cancer types. Nature. 579(7800):502–503. doi: 10.1038/d41586-020-00637-w.
- Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, Zhang Y, Gomez EC, Morreale B, Senchanthisai S, et al. 2022. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 40(2):153–167.e11. doi: 10.1016/j.ccell.2022.01.003.
- Alonso-Roman R, Last A, Mirhakkak MH, Sprague JL, Moller L, Grossmann P, Graf K, Gratz R, Mogavero S, Vylkova S, et al. 2022. Lactobacillus rhamnosus colonisation antagonizes Candida albicans by forcing metabolic adaptations that compromise pathogenicity. Nat Commun. 13(1):3192. doi: 10.1038/s41467-022-30661-5.
- Anderson JC, Robinson CM, Hisey WM, Edwards DK, Kneedler BL, Berger BM, Butterly LF. 2022. Colorectal neoplasia detection in individuals with positive multitarget stool DNA tests: Data from the New Hampshire colonoscopy registry. J Clin Gastroenterol. 56(5):419–425. doi: 10.1097/MCG.0000000000001554.
- Ashktorab H, Brim H. 2022. Colorectal cancer subtyping. Nat Rev Cancer. 22(2):68–69. doi: 10.1038/s41568-021-00432-3.
- Ballester MP, Mesonero F, Florez-Diez P, Gomez C, Fuentes-Valenzuela E, Martin N, Senosiain C, Vela M, Fernandez-Clotet A, Perez P, et al. 2022. Adherence to endoscopic surveillance for advanced lesions and colorectal cancer in inflammatory bowel disease: an AEG and GETECCU collaborative cohort study. Aliment Pharmacol Ther. 55(11):1402–1413. doi: 10.1111/apt.16832.
- Barbosa A, Miranda S, Azevedo NF, Cerqueira L, Azevedo AS. 2023. Imaging biofilms using fluorescence in situ hybridization: Seeing is believing. Front Cell Infect Microbiol. 13:1–16. doi: 10.3389/fcimb.2023.1195803.
- Bassetti M, Azoulay E, Kullberg BJ, Ruhnke M, Shoham S, Vazquez J, Giacobbe DR, Calandra T. 2021. EORTC/MSGERC definitions of invasive fungal diseases: Summary of activities of the intensive care unit working group. Clin Infect Dis. 72(Suppl 2):S121–S127. doi: 10.1093/cid/ciaa1751.
- Bilal H, Shafiq M, Hou B, Islam R, Khan MN, Khan RU, Zeng Y. 2022. Distribution and antifungal susceptibility pattern of Candida species from mainland China: A systematic analysis. Virulence. 13(1):1573–1589. doi: 10.1080/21505594.2022.2123325.
- Bing J, Wang S, Xu H, Fan S, Du H, Nobile CJ, Huang G. 2022. A case of Candida auris candidemia in Xiamen, China, and a comparative analysis of clinical isolates in China. Mycology. 13(1):68–75. doi: 10.1080/21501203.2021.1994479.
- Borman AM, Mohammed S, Palmer MD, Childs N, Johnson EM. 2022. The importance of appropriate processing and direct microscopic examination for the timely diagnosis and management of invasive infections caused by filamentous fungi. Med Mycol. 60(12):1–6. doi: 10.1093/mmy/myac081.
- Branco J, Miranda IM, Rodrigues AG. 2023. Candida parapsilosis virulence and antifungal resistance mechanisms: A comprehensive review of key determinants. J Fungi (Basel). 9(1):1–15. doi: 10.3390/jof9010080.
- Caesar LK, Butun FA, Robey MT, Ayon NJ, Gupta R, Dainko D, Bok JW, Nickles G, Stankey RJ, Johnson D, et al. 2023. Correlative metabologenomics of 110 fungi reveals metabolite-gene cluster pairs. Nat Chem Biol. 19(7):846–854. doi: 10.1038/s41589-023-01276-8.
- Cardoso R, Guo F, Heisser T, De Schutter H, Van Damme N, Nilbert MC, Tybjerg AJ, Bouvier AM, Bouvier V, Launoy G, et al. 2022. Proportion and stage distribution of screen-detected and non-screen-detected colorectal cancer in nine European countries: An international, population-based study. Lancet Gastroenterol Hepatol. 7(8):711–723. doi: 10.1016/S2468-1253(22)00084-X.
- Carethers JM, Doubeni CA. 2020. Causes of socioeconomic disparities in colorectal cancer and intervention framework and strategies. Gastroenterology. 158(2):354–367. doi: 10.1053/j.gastro.2019.10.029.
- Chang CW, Lee HC, Li LH, Chiang Chiau JS, Wang TE, Chuang WH, Chen MJ, Wang HY, Shih SC, Liu CY, et al. 2020. Fecal microbiota transplantation prevents intestinal injury, upregulation of toll-like receptors, and 5-fluorouracil/oxaliplatin-induced toxicity in colorectal cancer. Int J Mol Sci. 21(2):1–23. doi: 10.3390/ijms21020386.
- Cheung MK, Chan JYK, Wong MCS, Wong PY, Lei P, Cai L, Lan L, Ho WCS, Yeung ACM, Chan PKS, et al. 2022. Determinants and interactions of oral bacterial and fungal microbiota in healthy Chinese adults. Microbiol Spectr. 10(1):1–15. doi: 10.1128/spectrum.02410-21.
- Chung SS, Ali SI, Cash BD 2022. The present and future of colorectal cancer screening. Gastroenterol Hepatol (N Y). 18(11):646–653.
- Clay SL, Fonseca-Pereira D, Garrett WS. 2022. Colorectal cancer: The facts in the case of the microbiota. J Clin Invest. 132(4):1–10. doi: 10.1172/JCI155101.
- Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, Yu J. 2022. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. 10(1):1–12. doi: 10.1186/s40168-021-01208-5.
- Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J. 2019. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 68(4):654–662. doi: 10.1136/gutjnl-2018-317178.
- Collaborators GBDCC. 2022. Global, regional, and national burden of colorectal cancer and its risk factors, 1990-2019: A systematic analysis for the global burden of disease study 2019. Lancet Gastroenterol Hepatol. 7(7):627–647. doi: 10.1016/S2468-1253(22)00044-9.
- Dambuza IM, Brown GD. 2019. Fungi accelerate pancreatic cancer. Nature. 574(7777):184–185. doi: 10.1038/d41586-019-02892-y.
- Debeljak P, Baltar F. 2023. Fungal diversity and community composition across ecosystems. J Fungi (Basel). 9(5):1–10. doi: 10.3390/jof9050510.
- Dickson I. 2019. Fungal dysbiosis associated with colorectal cancer. Nat Rev Gastroenterol Hepatol. 16(2):76. doi: 10.1038/s41575-019-0105-2.
- Dohlman AB, Arguijo Mendoza D, Ding S, Gao M, Dressman H, Iliev ID, Lipkin SM, Shen X. 2021. The cancer microbiome atlas: A pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe. 29(2):281–298.e5. doi: 10.1016/j.chom.2020.12.001.
- Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, Iliev ID. 2022. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 185(20):3807–3822.e12. doi: 10.1016/j.cell.2022.09.015.
- Du H, Bing J, Nobile CJ, Huang G. 2022. Candida auris infections in China. Virulence. 13(1):589–591. doi: 10.1080/21505594.2022.2054120.
- Elmallah MIY, Ortega-Deballon P, Hermite L, Pais-De-Barros JP, Gobbo J, Garrido C. 2022. Lipidomic profiling of exosomes from colorectal cancer cells and patients reveals potential biomarkers. Mol Oncol. 16(14):2710–2718. doi: 10.1002/1878-0261.13223.
- Fassarella M, Blaak EE, Penders J, Nauta A, Smidt H, Zoetendal EG. 2021. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut. 70(3):595–605. doi: 10.1136/gutjnl-2020-321747.
- Foppiano Palacios C, Spichler Moffarah A. 2021. Diagnosis of pneumonia due to invasive molds. Diagnostics (Basel). 11(7):1–16. doi: 10.3390/diagnostics11071226.
- Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, Futran N, Houlton J, Sather C, Sicinska E, et al. 2022. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 611(7937):810–817. doi:10.1038/s41586-022-05435-0.
- Galindo-Pumarino C, Collado M, Herrera M, Pena C. 2021. Tumor microenvironment in metastatic colorectal cancer: The arbitrator in patients’ outcome. Cancers Basel. 13(5):1–28. doi: 10.3390/cancers13051130.
- Greathouse KL, Stone JK, Harris CC. 2020. Cancer-type-specific bacteria: Freeloaders or partners? Cancer Cell. 38(2):158–160. doi: 10.1016/j.ccell.2020.06.017.
- Guo R, Luo X, Xin X, Liu L, Wang X, Lu H. 2021. Microbial metabolomics: From methods to translational applications. Adv Exp Med Biol. 1280:97–113. doi: 10.1007/978-3-030-51652-9_7.
- Hindson J. 2019. Fungi promote pancreatic cancer. Nat Rev Gastroenterol Hepatol. 16(12):706–707. doi: 10.1038/s41575-019-0231-x.
- Hu LP, Huang W, Wang X, Xu C, Qin WT, Li D, Tian G, Li Q, Zhou Y, Chen S, et al. 2022. Terbinafine prevents colorectal cancer growth by inducing dNTP starvation and reducing immune suppression. Mol Ther. 30(10):3284–3299. doi: 10.1016/j.ymthe.2022.06.015.
- Huang J, Zheng X, Kang W, Hao H, Mao Y, Zhang H, Chen Y, Tan Y, He Y, Zhao W, et al. 2022. Metagenomic and metabolomic analyses reveal synergistic effects of fecal microbiota transplantation and anti-PD-1 therapy on treating colorectal cancer. Front Immunol. 13:1–11. doi: 10.3389/fimmu.2022.874922.
- Iliev ID, Cadwell K. 2021. Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology. 160(4):1050–1066. doi: 10.1053/j.gastro.2020.06.100.
- Joanito I, Wirapati P, Zhao N, Nawaz Z, Yeo G, Lee F, Eng CLP, Macalinao DC, Kahraman M, Srinivasan H, et al. 2022. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet. 54(7):963–975. doi: 10.1038/s41588-022-01100-4.
- Kamau E, Yang S. 2023. Metagenomic sequencing of positive blood culture fluid for accurate bacterial and fungal species identification: A pilot study. Microorganisms. 11(5):1–12. doi: 10.3390/microorganisms11051259.
- Kamel F, Eltarhoni K, Nisar P, Soloviev M. 2022. Colorectal cancer diagnosis: The obstacles we face in determining a non-invasive test and current advances in biomarker detection. Cancers Basel. 14(8):1–20. doi: 10.3390/cancers14081889.
- Kanth P, Inadomi JM. 2021. Screening and prevention of colorectal cancer. BMJ. 374(1855):n1855. doi: 10.1136/bmj.n1855.
- Kaur K, Zubair M, Adamski JJ. 2023. Fecal occult blood test. Treasure Island (FL): StatPearls Publishing.
- Ko KKK, Chng KR, Nagarajan N. 2022. Metagenomics-enabled microbial surveillance. Nature Microbiology. 7(4):486–496. doi: 10.1038/s41564-022-01089-w.
- Kong WL, Machida RJ. 2022. Development of transcriptomics-based growth rate indices in two model eukaryotes and relevance to metatranscriptomics datasets. Mol Ecol Resour. 22(7):2627–2639. doi: 10.1111/1755-0998.13652.
- Krawczyk A, Salamon D, Kowalska-Duplaga K, Zapala B, Ksiazek T, Drazniuk-Warchol M, Gosiewski T. 2023. Changes in the gut mycobiome in pediatric patients in relation to the clinical activity of crohn’s disease. World J Gastroenterol. 29(14):2172–2187. doi: 10.3748/wjg.v29.i14.2172.
- Kvaerner AS, Birkeland E, Bucher-Johannessen C, Vinberg E, Nordby JI, Kangas H, Bemanian V, Ellonen P, Botteri E, Natvig E, et al. 2021. The CRCbiome study: A large prospective cohort study examining the role of lifestyle and the gut microbiome in colorectal cancer screening participants. BMC Cancer. 21(1):1–14. doi: 10.1186/s12885-021-08640-8.
- Lam S, Bai X, Shkoporov AN, Park H, Wu X, Lan P, Zuo T. 2022. Roles of the gut virome and mycobiome in faecal microbiota transplantation. Lancet Gastroenterol Hepatol. 7(5):472–484. doi: 10.1016/S2468-1253(21)00303-4.
- Lapiere A, Richard ML. 2022. Bacterial-fungal metabolic interactions within the microbiota and their potential relevance in human health and disease: a short review. Gut Microbes. 14(1):e2105610. doi: 10.1080/19490976.2022.2105610.
- Lass-Florl C, Samardzic E, Knoll M. 2021. Serology anno 2021-fungal infections: From invasive to chronic. Clin Microbiol Infect. 27(9):1230–1241. doi: 10.1016/j.cmi.2021.02.005.
- Li C, Wang Y, Liu D, Wong CC, Coker OO, Zhang X, Liu C, Zhou Y, Liu Y, Kang W, et al. 2022. Squalene epoxidase drives cancer cell proliferation and promotes gut dysbiosis to accelerate colorectal carcinogenesis. Gut. 71(11):2253–2265. doi:10.1136/gutjnl-2021-325851.
- Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y, Chen H, Dai M. 2021. Incidence, mortality, survival, risk factor and screening of colorectal cancer: A comparison among China, Europe, and Northern America. Cancer Lett. 522:255–268. doi: 10.1016/j.canlet.2021.09.034.
- Li XV, Leonardi I, Putzel GG, Semon A, Fiers WD, Kusakabe T, Lin WY, Gao IH, Doron I, Gutierrez-Guerrero A, et al. 2022. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature. 603(7902):672–678. doi: 10.1038/s41586-022-04502-w.
- Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, Wong S, Szeto CH, Ng SC, Chan FKL, et al. 2020. A novel faecal lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 69(7):1248–1257. doi: 10.1136/gutjnl-2019-318532.
- Lin Y, Lau HC, Liu Y, Kang X, Wang Y, Ting NL, Kwong TN, Han J, Liu W, Liu C, et al. 2022. Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multicohort fecal metagenomic analyses. Gastroenterology. 163(4):908–921. doi:10.1053/j.gastro.2022.06.038.
- Lind AL, Pollard KS. 2021. Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome. 9(1):58. doi: 10.1186/s40168-021-01015-y.
- Liu C, Ng SK, Ding Y, Lin Y, Liu W, Wong SH, Sung JJ, Yu J. 2022. Meta-analysis of mucosal microbiota reveals universal microbial signatures and dysbiosis in gastric carcinogenesis. Oncogene. 41(28):3599–3610. doi: 10.1038/s41388-022-02377-9.
- Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang AJ, Chen J, Tao L, Zhou C, Fang W, et al. 2022. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat Microbiol. 7(2):238–250. doi:10.1038/s41564-021-01030-7.
- Liu X, Song Y, Li R. 2021. The use of combined PCR, fluorescence in situ hybridisation and immunohistochemical staining to diagnose mucormycosis from formalin-fixed paraffin-embedded tissues. Mycoses. 64(12):1460–1470. doi: 10.1111/myc.13382.
- Marongiu L, Allgayer H. 2022. Viruses in colorectal cancer. Mol Oncol. 16(7):1423–1450. doi: 10.1002/1878-0261.13100.
- Mauri G, Vitiello PP, Sogari A, Crisafulli G, Sartore-Bianchi A, Marsoni S, Siena S, Bardelli A. 2022. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br J Cancer. 127(3):394–407. doi: 10.1038/s41416-022-01769-8.
- McCarty TP, White CM, Pappas PG. 2021. Candidemia and invasive candidiasis. Infect Dis Clin North Am. 35(2):389–413. doi: 10.1016/j.idc.2021.03.007.
- Monahan KJ, Davies MM, Abulafi M, Banerjea A, Nicholson BD, Arasaradnam R, Barker N, Benton S, Booth R, Burling D, et al. 2022. Faecal immunochemical testing (FIT) in patients with signs or symptoms of suspected colorectal cancer (CRC): A joint guideline from the association of coloproctology of Great Britain and Ireland (ACPGBI) and the British society of Gastroenterology (BSG). Gut. 71(10):1939–1962. doi: 10.1136/gutjnl-2022-327985.
- Moreira FM, Pereira PA, Miranda R, Reis C, Braga L, de Andrade JM, Do Nascimento LG, Mattoso JMV, Forsythe SJ, da Costa LV, et al. 2023. Evaluation of MALDI-TOF MS, sequencing of D2 LSU rRNA and internal transcribed spacer regions (ITS) for the identification of filamentous fungi isolated from a pharmaceutical facility. J Pharm Biomed Anal. 234:115531. doi: 10.1016/j.jpba.2023.115531.
- Mukherjee C, Leys EJ. 2021. Strain-level profiling of oral microbiota with targeted sequencing. Methods Mol Biol. 2327:239–252. doi: 10.1007/978-1-0716-1518-8_14.
- Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, Gavert N, Stajich JE, Amit G, Gonzalez A, et al. 2022. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. 185(20):3789–3806.e17. doi: 10.1016/j.cell.2022.09.005.
- Nassar FJ, Msheik ZS, Nasr RR, Temraz SN. 2021. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin Epigenet. 13(1):111. doi: 10.1186/s13148-021-01095-5.
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. 2020. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Sci. 368(6494):973–980. doi: 10.1126/science.aay9189.
- Nel Van Zyl K, Whitelaw AC, Hesseling AC, Seddon JA, Demers AM, Newton-Foot M. 2022. Fungal diversity in the gut microbiome of young South African children. BMC Microbiol. 22(1):1–11. doi: 10.1186/s12866-022-02615-w.
- Noh JH, Shin JY, Lee JH, Park YS, Lee IS, Kim GH, Na HK, Ahn JY, Jung KW, Kim DH, et al. 2023. Clinical significance of Epstein-Barr virus and Helicobacter pylori infection in gastric carcinoma. Gut Liver. 17(1):69–77. doi: 10.5009/gnl210593.
- O’Sullivan DE, Sutherland RL, Town S, Chow K, Fan J, Forbes N, Heitman SJ, Hilsden RJ, Brenner DR. 2022. Risk factors for early-onset colorectal cancer: A systematic review and meta-analysis. Clin Gastroenterol Hepatol. 20(6):1229–1240.e5. doi: 10.1016/j.cgh.2021.01.037.
- Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. 2022. The rising tide of early-onset colorectal cancer: A comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol. 7(3):262–274. doi: 10.1016/S2468-1253(21)00426-X.
- Perez JC. 2021. The interplay between gut bacteria and the yeast Candida albicans. Gut Microbes. 13(1):e1979877. doi: 10.1080/19490976.2021.1979877.
- Petriglieri F, Singleton CM, Kondrotaite Z, Dueholm MKD, McDaniel EA, McMahon KD, Nielsen PH, McGrath J. 2022. Reevaluation of the phylogenetic diversity and global distribution of the genus “Candidatus accumulibacter”. mSystems. 7(3):1–15. doi: 10.1128/msystems.00016-22.
- Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, et al. 2020. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 579(7800):567–574. doi: 10.1038/s41586-020-2095-1.
- Pratt M, Forbes JD, Knox NC, Van Domselaar G, Bernstein CN. 2022. Colorectal cancer screening in inflammatory bowel diseases—Can characterization of GI microbiome signatures enhance Neoplasia detection? Gastroenterology. 162(5):1409–1423.e1. doi: 10.1053/j.gastro.2021.12.287.
- Qin Y, Havulinna AS, Liu Y, Jousilahti P, Ritchie SC, Tokolyi A, Sanders JG, Valsta L, Brozynska M, Zhu Q, et al. 2022. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat Genet. 54(2):134–142. doi: 10.1038/s41588-021-00991-z.
- Qu R, Ma Y, Zhang Z, Fu W. 2022. Increasing burden of colorectal cancer in China. Lancet Gastroenterol Hepatol. 7(8):700. doi: 10.1016/S2468-1253(22)00156-X.
- Randel KR, Schult AL, Botteri E, Hoff G, Bretthauer M, Ursin G, Natvig E, Berstad P, Jorgensen A, Sandvei PK, et al. 2021. Colorectal cancer screening with repeated fecal immunochemical test versus sigmoidoscopy: Baseline results from a randomized trial. Gastroenterology. 160(4):1085–1096.e5. doi: 10.1053/j.gastro.2020.11.037.
- Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S. 2021. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 591(7851):633–638. doi: 10.1038/s41586-021-03241-8.
- Rayens E, Norris KA, Cordero JF. 2022. Mortality trends in risk conditions and invasive mycotic disease in the United States, 1999-2018. Clin Infect Dis. 74(2):309–318. doi: 10.1093/cid/ciab336.
- Rebersek M. 2021. Gut microbiome and its role in colorectal cancer. BMC Cancer. 21(1):1325. doi: 10.1186/s12885-021-09054-2.
- Sampaio KB, Dos Santos Nascimento D, Garcia EF, de Souza EL. 2022. An outlook on fluorescent in situ hybridization coupled to flow cytometry as a versatile technique to evaluate the effects of foods and dietary interventions on gut microbiota. Arch Microbiol. 204(8):469. doi: 10.1007/s00203-022-03090-7.
- Sarantis P, Trifylli EM, Koustas E, Papavassiliou KA, Karamouzis MV, Papavassiliou AG. 2022. Immune microenvironment and immunotherapeutic management in virus-associated digestive system tumors. Int J Mol Sci. 23(21):1–20. doi: 10.3390/ijms232113612.
- Schmitt M, Greten FR. 2021. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 21(10):653–667. doi: 10.1038/s41577-021-00534-x.
- Shah SC, Itzkowitz SH. 2022. Colorectal cancer in inflammatory bowel disease: Mechanisms and management. Gastroenterology. 162(3):715–730.e3. doi: 10.1053/j.gastro.2021.10.035.
- Shen S, Huo D, Ma C, Jiang S, Zhang J, Xu ZZ. 2021. Expanding the colorectal cancer biomarkers based on the human gut phageome. Microbiol Spectr. 9(3):e0009021. doi: 10.1128/Spectrum.00090-21.
- Shi L, Xu Y, Feng M. 2023. Role of gut microbiome in immune regulation and immune checkpoint therapy of colorectal cancer. Dig Dis Sci. 68(2):370–379. doi: 10.1007/s10620-022-07689-0.
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. 2020. Gastric cancer. Lancet. 396(10251):635–648. doi: 10.1016/S0140-6736(20)31288-5.
- Stasiewicz M, Karpinski TM. 2022. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 86(Pt 3):633–642. doi: 10.1016/j.semcancer.2021.11.002.
- Stewart OA, Wu F, Chen Y. 2020. The role of gastric microbiota in gastric cancer. Gut Microbes. 11(5):1220–1230. doi: 10.1080/19490976.2020.1762520.
- Strickland AB, Shi M. 2021. Mechanisms of fungal dissemination. Cell Mol Life Sci. 78(7):3219–3238. doi: 10.1007/s00018-020-03736-z.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A Cancer J Clin. 71(3):209–249. doi: 10.3322/caac.21660.
- Sung SY, Choi HH, Kim S, Park BR, Kim YK, Kim HK, Cho YS, Kim SW, Kim SS, Chae HS. 2022. Colonoscopy decreases mortality in colorectal cancer patients compared with fecal immunochemical test. J Gastroenterol Hepatol. 37(10):1991–1997. doi: 10.1111/jgh.15924.
- Tang C, Sun H, Kadoki M, Han W, Ye X, Makusheva Y, Deng J, Feng B, Qiu D, Tan Y, et al. 2023. Blocking Dectin-1 prevents colorectal tumorigenesis by suppressing prostaglandin E2 production in myeloid-derived suppressor cells and enhancing IL-22 binding protein expression. Nat Commun. 14(1):1493. doi:10.1038/s41467-023-37229-x.
- Vehreschild JJ, Koehler P, Lamoth F, Prattes J, Rieger C, Rijnders BJA, Teschner D. 2021. Future challenges and chances in the diagnosis and management of invasive mould infections in cancer patients. Med Mycol. 59(1):93–101. doi: 10.1093/mmy/myaa079.
- Wang H, Wu H, Li KD, Wang YY, Huang RG, Du YJ, Jin X, Zhang QR, Li XB, Li BZ. 2023. Intestinal fungi and systemic autoimmune diseases. Autoimmun Rev. 22(2):103234. doi: 10.1016/j.autrev.2022.103234.
- Wang N, Fang JY. 2023. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31(2):159–172. doi: 10.1016/j.tim.2022.08.010.
- Wang S, Zhang YR, Yu YB. 2021. The important role of fungi in inflammatory bowel diseases. Scand J Gastroenterol. 56(11):1312–1322. doi: 10.1080/00365521.2021.1963838.
- Xiao Q, Lu W, Kong X, Shao YW, Hu Y, Wang A, Bao H, Cao R, Liu K, Wang X, et al. 2021. Alterations of circulating bacterial DNA in colorectal cancer and adenoma: A proof-of-concept study. Cancer Lett. 499:201–208. doi: 10.1016/j.canlet.2020.11.030.
- Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, Wang F, Yan H, Zeng L, Zhang L, et al. 2022. Microbiota in tumors: From understanding to application. Adv Sci. 9(21):e2200470. doi:10.1002/advs.202200470.
- Xu H, Chen H, Hu J, Xiong Z, Li D, Wang S, Yu J. 2022. Feasibility of quantification based on novel evaluation with stool DNA and fecal immunochemical test for colorectal cancer detection. BMC Gastroenterol. 22(1):384. doi: 10.1186/s12876-022-02470-z.
- Xu J. 2021. Is natural population of Candida tropicalis sexual, parasexual, and/or asexual? Front Cell Infect Microbiol. 11:751676. doi: 10.3389/fcimb.2021.751676.
- Xu J, Ren X, Liu Y, Zhang Y, Zhang Y, Chen G, Huang Q, Liu Q, Zhou J, Liu Y. 2022. Alterations of fungal microbiota in patients with cholecystectomy. Front Microbiol. 13:831947. doi: 10.3389/fmicb.2022.831947.
- Yelorda KL, Fu SJ, Owens DK. 2021. Analysis of survival among adults with early-onset colorectal cancer. JAMA Netw Open. 4(6):e2112878. doi: 10.1001/jamanetworkopen.2021.12878.
- Yu H, Li XX, Han X, Chen BX, Zhang XH, Gao S, Xu DQ, Wang Y, Gao ZK, Yu L, et al. 2023. Fecal microbiota transplantation inhibits colorectal cancer progression: Reversing intestinal microbial dysbiosis to enhance anti-cancer immune responses. Front Microbiol. 14:1126808. doi: 10.3389/fmicb.2023.1126808.
- Yu M, Ding H, Gong S, Luo Y, Lin H, Mu Y, Li H, Li X, Zhong M. 2023. Fungal dysbiosis facilitates inflammatory bowel disease by enhancing CD4+ T cell glutaminolysis. Front Cell Infect Microbiol. 13:1140757. doi: 10.3389/fcimb.2023.1140757.
- Yuan B, Ma B, Yu J, Meng Q, Du T, Li H, Zhu Y, Sun Z, Ma S, Song C. 2021. Fecal bacteria as non-invasive biomarkers for colorectal adenocarcinoma. Front Oncol. 11:664321. doi: 10.3389/fonc.2021.664321.
- Zajta E, Csonka K, Toth A, Tiszlavicz L, Nemeth T, Orosz A, Novak A, Csikos M, Vagvolgyi C, Mocsai A, et al. 2021. Signaling through Syk or CARD9 mediates species-specific anti-Candida protection in bone marrow chimeric mice. mBio. 12(4):e0160821. doi: 10.1128/mBio.01608-21.
- Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S, Zhu Y, Miao J, Xiong S, Fei S, et al. 2020. Aberrant DNA methylation of SEPT9 and SDC2 in stool specimens as an integrated biomarker for colorectal cancer early detection. Front Genet. 11:643. doi: 10.3389/fgene.2020.00643.
- Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, Wang H, Gu D, Zhu L, Li S, et al. 2022. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIf3h interaction. Mol Cancer. 21(1):49. doi: 10.1186/s12943-021-01471-y.
- Zheng X, Wang R, Zhang X, Sun Y, Zhang H, Zhao Z, Zheng Y, Luo J, Zhang J, Wu H, et al. 2022. A deep learning model and human-machine fusion for prediction of EBV-associated gastric cancer from histopathology. Nat Commun. 13(1):2790. doi: 10.1038/s41467-022-30459-5.
- Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu Z, Song Y, Hong X. 2021. Candida albicans disorder is associated with gastric carcinogenesis. Theranostics. 11(10):4945–4956. doi: 10.7150/thno.55209.
- Zhou H, Zhu L, Song J, Wang G, Li P, Li W, Luo P, Sun X, Wu J, Liu Y, et al. 2022. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer. 21(1):86. doi: 10.1186/s12943-022-01556-2.
- Zhu Y, Shi T, Lu X, Xu Z, Qu J, Zhang Z, Shi G, Shen S, Hou Y, Chen Y, et al. 2021. Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J. 40(11):e105320. doi: 10.15252/embj.2020105320.
- Zozaya-Valdes E, Wong SQ, Raleigh J, Hatzimihalis A, Ftouni S, Papenfuss AT, Sandhu S, Dawson MA, Dawson SJ. 2021. Detection of cell-free microbial DNA using a contaminant-controlled analysis framework. Genome Biol. 22(1):187. doi: 10.1186/s13059-021-02401-3.