ABSTRACT
Candida albicans is one of the most common opportunistic fungi in cancer patients. This study explored the influence of C. albicans on gut microbiota in oral tumour-bearing mice by means of 16S rRNA sequencing and ITS sequencing. It was found that C. albicans infection induced the decrease of alpha diversity of bacteria and fungi in the gut microbiome. For the bacteria, C. albicans caused the reduction of Ralstonia, Alistipes, Clostridia UCG-014, Ruminococcus, and Lachnospiraceae NK4A136 group. For the fungi, C. albicans inhibited the growth of other fungi including Aspergillus, Cladosporium, and Bipolaris. The neutralisation of γδT cells partly alleviated the out-of-balance of Firmicutes/Bacteroidota (F/B) ratio in the gut caused by C. albicans infection. However, γδT cell neutralisation boosted the overgrowth of C. albicans. Additionally, IL-17A neutralisation aggravated the microbial dysbiosis of bacteria and fungi caused by C. albicans infection. Further analysis indicated that C. albicans overgrowth might influence the correlations between fungal and bacterial kingdoms. In conclusion, C. albicans infection disturbed the gut microbiota of both bacteria and fungi in oral tumour-bearing mice, which may be associated with the intestinal immune components including γδT cells and IL-17A.
1. Introduction
Candida albicans (C. albicans) is the most common opportunistic fungus inhabiting in the human oral cavity and intestinal tract, which shows close associations with various types of cancer including oral cancer (OC), oesophageal cancer, liver cancer, and colorectal cancer (Wang et al. Citation2023b). Among them, it was estimated that C. albicans infection could be detected in approximately 10%–68.2% of OC patients (Wang et al. Citation2023b).
OC is the malignant neoplasm of the lip and oral cavity. Candida presence is not only a risk factor for OC development but also associated with poor overall survival in OC patients (Mohamed et al. Citation2021). Both the cancer itself and cancer treatments including chemotherapy and radiotherapy are contributors to the overgrowth of C. albicans (Panghal et al. Citation2012). Unfortunately, C. albicans infection may induce mucosal bacterial dysbiosis (Bertolini et al. Citation2019; Zaongo et al. Citation2023), while gut microbiome influences immunotherapy against epithelial tumours (Routy et al. Citation2018b). However, little is known about the influence of C. albicans overgrowth during OC on the gut microbiota.
The human microbiota is a dynamic set of 40 trillion microbes, which is made up of more than 3,000 species containing bacteria, fungi, and viruses (Ting et al. Citation2022). The inter-kingdom interactions between bacteria and fungi are necessary for the development of the host immune system and homoeostasis of the mucosal barrier (Takiishi et al. Citation2017). The dyshomeostasis of microbiota is not only a reflection of pathological status, but may have a significant effect on host health and tumour therapeutic outcomes (Gopalakrishnan et al. Citation2018; Helmink et al. Citation2019).
Both the commensal microbiota and immune system contribute to the homoeostasis of the intestinal mucosal barrier. A previous study has revealed that C. albicans infection was associated with the loss of mucosal bacterial diversity in intestinal mucosa in mice receiving 5-fluorouracil (Bertolini et al. Citation2019). In turn, the commensal intestinal bacteria may protect the host from C. albicans challenge by altering their species diversity (Wang et al. Citation2021). However, whether immune factors play roles in the microbial disturbance caused by C. albicans infection is unclear.
Among the various types of immune cells and cytokines, γδT cells and IL-17A are important factors in response to C. albicans infection. γδT cells make up 10%–30% of CD3+ T cells in the human intestine, which play important roles in maintaining intestinal homoeostasis and resisting invasive pathogens (Shiromizu and Jancic Citation2018). During immune response, γδT cells usually act earlier than αβT cells and serve as a bridge between innate and adaptive immunity. In the early stage of the innate immune response, γδT cells recruit innate cells including neutrophils and macrophages; then in the middle stage, they regulate B cells to produce immunoglobulins, and present antigens to CD4+ T cells and CD8+ T cells; however, in the later stage, they can kill macrophages and αβT cells and promote tissue repair (Zhou et al. Citation2020). Additionally, IL-17A is an important cytokine during C. albicans infection, which is mainly produced by Th17 cells and γδT cells (Majumder and Mcgeachy Citation2021). The bidirectional interactions between γδT cells or IL-17A and microbiota have been noticed but not been fully understood (Majumder and Mcgeachy Citation2021; Papotto et al. Citation2021). Additionally, whether γδT cells and IL-17A play roles in influencing gut microbiota during C. albicans overgrowth has not been determined.
Considering the overgrowth of C. albicans in OC patients, the unrevealed interplay between C. albicans infection and gut microbes, as well as the undetermined roles of γδT cells and IL-17A in C. albicans-gut microbiota interplay, this study aimed to explore the changes of gut microbiome caused by C. albicans overgrowth in OC mice and the role of γδT cells and IL-17A during this process. As a result, this study exhibited the significant influence of C. albicans overgrowth on gut bacteria and fungi in oral tumour-bearing mice. Additionally, it was revealed that immune components (IL-17A and γδT cells) might play potential roles in modulating gut microbiota during C. albicans overgrowth.
2. Materials and methods
2.1. Oral tumour-bearing mouse model
C3H/HeN mice (6 to 8 weeks) were used in the study. All mice were maintained under specific pathogen-free conditions, fed with the same feed, and in the same facility and housing unit. Fresh faeces in the colon of oral tumour-bearing mice were collected with sterile instruments from our previous work (Wang et al. Citation2023a), and stored at −80 °C. Briefly, C3H/HeN mice were fed with 0.1% (wt/vol) tetracycline hydrochloride in drinking water for 1 week. Then, the mice in C. albicans infected group (CA group) were infected with C. albicans (SC5314 strain) by drinking water containing C. albicans for 2 weeks. After 2 weeks of pre-infection, all of the infected (CA group) and uninfected (CON group) mice were injected with SCC VII cells into the submucosa of the tongue dorsum to induce oral tumour. Two weeks later, the faeces were collected. The animal experiments were approved by the Biomedical Ethics Committee of Peking University.
2.2. In vivo neutralisation of γδT cells or IL-17A
The in vivo neutralisation experiments were performed as previously (Wang et al. Citation2023a). As for the IL-17A neutralising experiment, the oral tumour-bearing mice were injected both intraperitoneally (50 μg, 2 times/week) and intratumorally (25 μg, 3 times/week) with anti-mouse IL-17A antibody (clone 17F3, BioXcell) for 2 weeks (AIL group: Mice with C. albicans infection plus IL-17A neutralisation). As for the γδT cell neutralising experiment, the tumour-bearing mice were also injected both intraperitoneally (50 μg, 2 times/week) and intratumorally (25 μg, 3 times/week) with anti-mouse TCRγ/δ antibody (clone UC7-13D5, BioXcell) for 2 weeks (ATG group: Mice with C. albicans infection plus TCRγ/δ neutralisation).
2.3. 16S rRNA and ITS sequencing
The 16S rRNA and ITS sequencings were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Briefly, DNA was extracted from faecal pellets using a Magnetic Soil and Stool DNA kit (TianGen, China, Catalog #: DP712). The DNA was amplified using 515F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806 R (5’-GGACTACHVGGGTWTCTAAT-3’) to target the V4 region of bacteria or using ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3’) to target the ITS region of fungi. Then the samples were barcoded and pooled to construct the sequencing library, and sequenced by an Illumina Novaseq6000 to generate pair-ended 150 × 150 reads.
2.4. Bioinformatics analysis
Paired-end reads were merged using FLASH (V1.2.11) (Magoc and Salzberg Citation2021). Fastp (V0.23.1) software was used to perform quality filtering and obtain high-quality Clean Tags. The clean tags were compared with the reference database [Silva database (for 16S), https://www.arb-silva.de/; Unite Database (for ITS), https://unite.ut.ee/] using UCHIME Algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html) to remove the chimera sequences (Edgar et al. Citation2011). Then, the effective tags were used to obtain ASVs (Amplicon Sequence Variants) with DADA2 or deblur module in the QIIME2 software (Version QIIME2–202006). Species annotation was performed using QIIME2 software (16S: Silva Database, ITS: Unite Database). Multiple sequencing alignment was performed using QIIME2 software. Then, the absolute abundance of ASVs was normalised using a standard sequence number corresponding to the sample with the least sequences. Subsequent analysis of alpha diversity and beta diversity were all performed based on the output normalised data. The correlation analyses between 16S and ITS data was performed using Omicsmart, a dynamic real-time interactive online platform for data analysis (http://www.omicsmart.com).
2.5. Statistics
For the differential analysis of Chao1 richness, Observed OTUs, Shannon, and Pielou evenness index, a Student’s t-test was used to determine the statistical relevance between two groups. Bray-Curtis distance was used to analyse the beta-diversity patterns. T-test was used to find out the differential species between groups. Wilcoxon test was used to analyse the Bray-Curtis distance between inter- and intra-group. Procrustes test was used to analyse the correlation between 16S and ITS in groups. Spearman correlation analysis was used to analyse the interactions between 16S and ITS data. Values of P < 0.05 were considered significant.
2.6. Data availability
Sequence data that support the findings of this study have been deposited in NCBI BioProject with the primary accession codes (PRJNA991314 for 16S, PRJNA991317 for ITS) and in NCBI Sequence Read Archive (SRA) with the primary accession codes (SRP447344 for 16S, SRP447374 for ITS).
3. Results
3.1. C. albicans infection disturbed the gut microbiota of oral tumour-bearing mice
In the 16S rRNA sequencing, a rarefaction curve based on the observed OTUs showed that the sequencing depth was sufficient to detect species in the samples (). Alpha diversity was estimated by Chao1, Observed OTUs, Pielou evenness, and Shannon index (). The results showed that the Observed OTUs (P = 0.015), Pielou evenness (P = 0.005), and Shannon index (P = 0.002) in the CON group were significantly higher than in the CA group (), which indicated that the C. albicans infection caused the reduction of community richness and diversity of gut bacteria in tumour-bearing mice. Beta diversity estimated by the PCoA plot showed a distinguishable microbial composition between the CON and CA groups ().
Figure 1. Candida albicans overgrowth induced the disorder of the gut microbiota of bacteria.
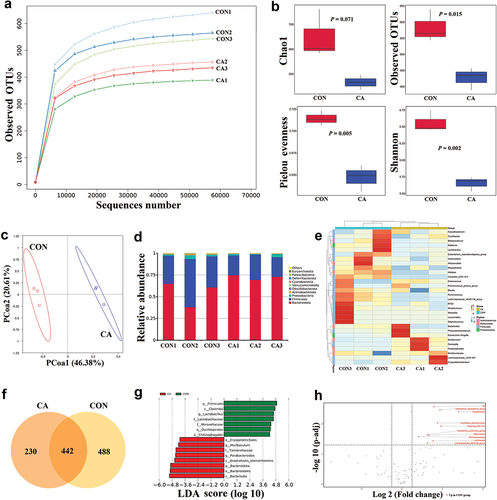
To further study the changes in the gut bacteria between the CON and CA groups, the compositional changes were evaluated. At the phylum level, it was noticed that the Bacteroidota was increased, but Firmicutes was reduced in the CA group compared to the CON group (). At the genus level, it was found that Ralstonia, Alistipes, Clostridia UCG-014, Ruminococcus, and Lachnospiraceae NK4A136 group were reduced, but Muribaculum, Dubosiella, Parabacteroides, Lachnospiraceae UCG-001, and Erysipelatoclostridium were increased in the CA group (). The Venn graph showed that there were 442 common ASVs between CON and CA groups, 488 unique ASVs in the CON group, and 230 unique ASVs in the CA group (). Further, the LEfSe (LDA Effect Size) result showed that Firmicutes, Clostridia, Lactobacillus, et al. were the main taxa enriched in the CON group, while Bacteroidales, Muribaculum, Parabacteroides, et al. were more abundant in the CA group (LDA score > 4, P < 0.05) (). The volcano plot showed the differentially altered species between groups, which indicated that C. albicans caused the significant reduction of various bacteria, such as Clostridia UCG-014, Akkermansia, and Lachnospira in the gut of mice (P < 0.05) ().
In the ITS gene sequencing, the sequencing depth was also sufficient to detect species in the samples (). The alpha diversity showed that the Pielou evenness (P = 0.000779) and Shannon (P = 0.0168) index in the CON group were significantly higher than in the CA group (). The PCoA plot showed a distinguishable fungal composition between the CON and CA groups ().
Figure 2. Candida albicans overgrowth induced the disorder of gut microbiota of fungi.
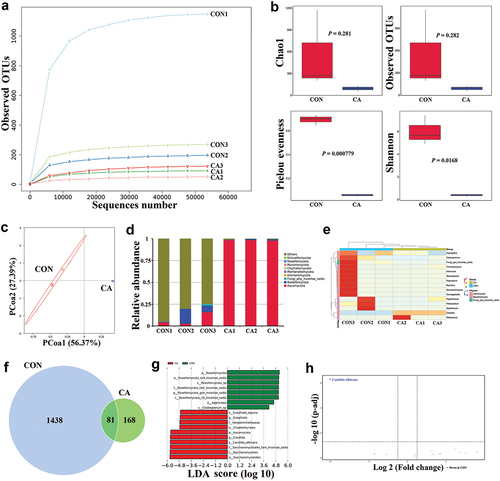
The compositional changes evaluated at the phylum level showed that Ascomycota was significantly increased in the CA group compared to the CON group (). At the genus level, it was found that Candida and Malassezia were increased, but others including Aspergillus, Cladosporium, and Bipolaris were reduced in the CA group (). The Venn graph showed that there were 81 common ASVs between the CON and CA groups, 1,438 unique ASVs in the CON group and only 168 unique ASVs in the CA group (). Further, the LEfSe result showed that Rozellomycota, Agaricales, and Cladosporium were the main taxa enriched in the CON group, while Exophiala and Candida were more abundant in the CA group (LDA score > 4, P < 0.05) (). The volcano plot demonstrated that C. albicans was the dominant fungus in the gut of mice with C. albicans infection () (P < 0.05).
3.2. In vivo γδT cell neutralisation partly rescued the bacterial dysbiosis, but aggravated the fungal dysbiosis caused by C. albicans infection
γδT cells are a cluster of cells much more abundant as tissue-resident intraepithelial lymphocytes in the gastrointestinal tract (Ivanov et al. Citation2022), which is one of the important parts during Candida infection (Mengesha and Conti Citation2017). To explore the role of γδT cells in the microecological disturbance caused by C. albicans infection, an in vivo γδT cell neutralisation experiment was performed.
As for 16S sequencing, the rarefaction curve showed that the sequencing depth was sufficient to detect species in the samples (). The alpha diversity showed that the Chao1, Observed OTUs, Pielou evenness, and Shannon index in the ATG group were higher than in the CA group, though the differences were not statistically significant (P > 0.05) (). The PCoA plot indicated that there was no significant difference in beta diversity between the CA and ATG groups ().
Figure 3. In vivo γδT cell neutralisation partly rescued the bacterial dysbiosis but aggravated the fungal dysbiosis caused by Candida albicans overgrowth.
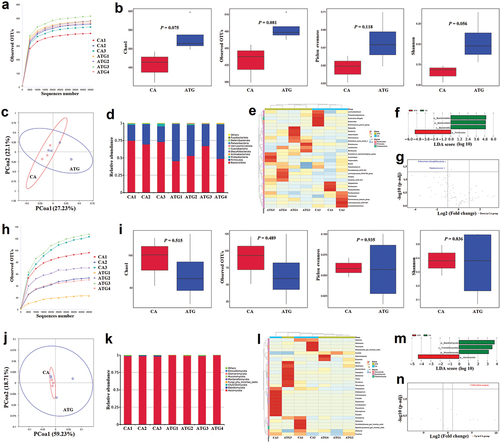
When compared to the CA group, the Bacteroidota was decreased, but Firmicutes was increased in the ATG group (), which indicated that the neutralisation of γδT cells partly alleviated the out-of-balance of Firmicutes/Bacteroidota (F/B) ratio in the gut caused by C. albicans infection. At the genus level, it was found that the reduced bacteria including Alistipes, Clostridia UCG-014, Alloprevotella, and Lachnospiraceae NK4A136 group caused by C. albicans infection were partly rescued after the neutralisation of γδT cells (). The LEfSe result showed that Firmicutes was the main taxa enriched in the ATG group, while Bacteroidota was more abundant in the CA group (LDA score > 3, P < 0.05) (). Additionally, it was shown that Eubacterium xylanophilum group and Ruminococcaceae increased in the ATG group when compared to the CA group (P < 0.05) ().
As for the ITS sequencing, the sequencing depth was also sufficient to detect species in the samples (). The alpha diversity showed that the Chao1 and Observed OTUs index in the ATG group were lower than in the CA group, though there were no statistically significant differences (P > 0.05) (). The PCoA plot indicated that there was no difference in beta diversity between the CA and ATG groups ().
The Basidiomycota in the ATG group was lower than in the CA group (). At the genus level, it was found that Wallemia and Filobasidium were decreased but Candida and Exophiala were increased in the ATG group compared to the CA group (). The LEfSe result showed that Ascomycota was more abundant in the ATG group compared to the CA group (LDA score > 4, P < 0.05) (). Additionally, it was shown that Filobasidium magnum was significantly decreased in the ATG group (P < 0.05) ().
3.3. In vivo IL-17A neutralisation aggravated the microbial dysbiosis caused by C. albicans infection
IL-17A is an important cytokine for antifungal immunity, which mediates the antimicrobial immune response and contributes to wound healing of injured epithelium (Akuzum and Lee Citation2022). Thus, we further explored the influence of IL-17A on gut microbiota during C. albicans infection.
The sequencing depth was sufficient to detect species in the samples (). The alpha diversity of bacteria showed that the Chao1 and Observed OTUs index in the AIL group were lower than in the CA group, though the differences were not statistically significant (P > 0.05) (). The PCoA plot indicated that there was no significant difference in beta diversity between the CA and AIL groups ().
Figure 4. In vivo IL-17A neutralisation aggravated the microbial dysbiosis caused by Candida albicans overgrowth.
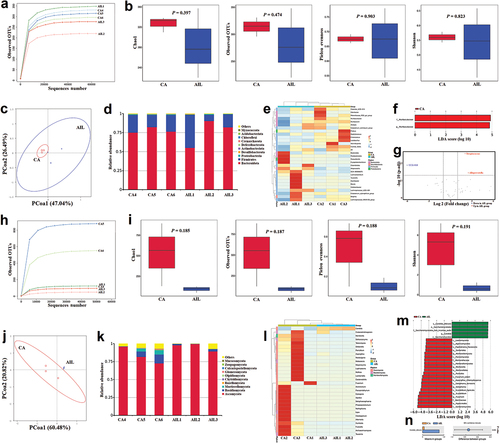
When compared to the CA group, the Bacteroidota was increased, but Firmicutes was decreased in the AIL group (), which indicated that the neutralisation of IL-17A aggravated the out-of-balance of F/B in the gut caused by C. albicans infection. At the genus level, it was found that the neutralisation of IL-17A reduced Clostridia UCG-014, Alistipes, Muribaculum, Straphylococcus, Enterococcus, and Lactobacillus, but increased Alloprevotella, GCA-900066575, and Lachnoclostridium in mice gut with C. albicans infection (). Additionally, it was found that Muribaculaceae was the main taxa enriched in the CA group (). The volcano plot demonstrated that the neutralisation of IL-17A reduced the UCG-010, but increased the Alloprevotella and Streptococcus in the gut of mice with C. albicans infection (P < 0.05) ().
As for the mycobiome, it was found that the Chao1, Observed OTUs, Pielou evenness, and Shannon index in the AIL group were lower than in the CA group, though there were no significant differences (P > 0.05) (). The PCoA plot showed a difference in beta diversity between the CA and AIL groups (). When compared to the CA group, the Ascomycota was increased, but other fungi were decreased in the AIL group (), which indicated that the neutralisation of IL-17A aggravated the fungal disorder caused by C. albicans infection. It was found that the neutralisation of IL-17A boosted the C. albicans overgrowth (P < 0.05) but inhibited most of the other fungi in the gut ().
3.4. Significant correlations between fungal and bacterial kingdoms were found in the gut microbiome during C. albicans infection
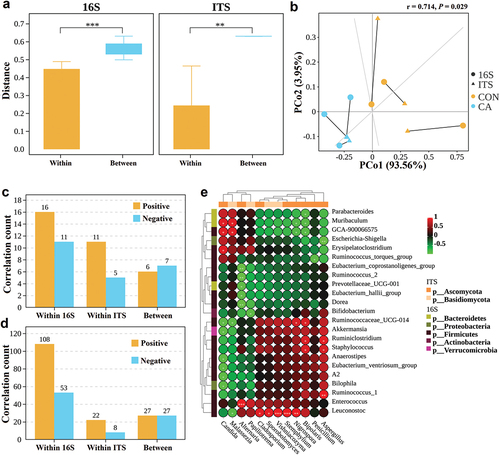
Pairwise comparisons of Bray-Curtis dissimilarity values (Nash et al. Citation2017) based on OTUs between samples from the same groups (within the CON group or the CA group) and between samples from different groups (between CON and CA group) for the 16S rRNA and ITS sequencing data were performed. The results showed that the Bray-Curtis distance for both 16S and ITS was significantly higher in inter-group (between) than in intra-group (within) (P < 0.01) (), which indicated that the C. albicans infection influenced both the bacterial and fungal microbiome. Procrustes correlation testing (Longa et al. Citation2017) for PCoA analysis showed a correlation of 0.714 at Class level (M2 = 0.4896, P < 0.05) (), meaning that bacterial and fungal diversity reacted in a similar way to C. albicans challenge. Then, the correlations between fungal and bacterial kingdoms were analysed. In the family level, the proportion of inter-kingdom similarity values with an absolute value > 0.5 (P < 0.05) that were positive was 46.2% (6 positives, 7 negatives) (). In the genus level, the proportion of positive intra-kingdom bacterial and fungal similarity values was 67.1% (108 positives, 53 negatives) and 73.3% (22 positives, 8 negatives), respectively, both of which were higher than the proportion of inter-kingdom positive correlations (P = 0.037 and 0.065 respectively) (). Comparing abundances of fungal and bacterial genera revealed that there were strong positive correlations between Candida and Muribaculum, GCA-900066575, and Erysipelatoclostridium, but negative correlations between Candida and Ruminococcaceae UCG-014, Akkermansia, Anaerostipes, Eubacterium ventriosum group, A2, and Bilophila ().
Figure 5. Significant correlations between fungal and bacterial kingdoms were found in the gut microbiome during Candida albicans overgrowth.
4. Discussions
C. albicans is the main pathogen to oropharyngeal and gastrointestinal candidiasis in cancer patients receiving chemotherapy and radiotherapy (Umazume et al. Citation1995; Bertolini et al. Citation2019; Kumari et al. Citation2021). In this study, we investigated the influence of C. albicans infection on the composition of intestinal bacteria and fungi in the context of oral cancer. It was demonstrated that C. albicans overgrowth led to a profound taxonomic imbalance on the gut microbiota. It was also discovered that the neutralisation of γδT cells partly rescued the bacterial dysbiosis caused by C. albicans infection, while the neutralisation of IL-17A aggravated the microbial dysbiosis caused by C. albicans infection. Additionally, the inter-kingdom relationships between bacteria and fungi seemed to be influenced during C. albicans overgrowth. Thus, the results indicated that the overgrowth of C. albicans in oral cancer patients could induce the dysbiosis of gut microbiota, which was partly associated with the intestinal immune components including γδT cells and IL-17A.
In the present study, C. albicans infection disrupted the gut microbiota including bacteria and fungi in oral tumour-bearing mice. As for bacteria, the alpha diversity and F/B ratio were significantly reduced after C. albicans infection. Firmicutes and Bacteroidota are two dominant phyla occupying together up to 90% of the total gut microbiota (Human Microbiome Project Consortium Citation2012). The F/B ratio is suggested to be an important index of the health of gut microbiota (Vaiserman et al. Citation2020). The reduced gut F/B ratio is associated with many pathological conditions including breast cancer (An et al. Citation2023), Hepatitis C virus (Aly et al. Citation2016), and inflammatory bowel disease (Stojanov et al. Citation2020). Thus, the reduced F/B ratio caused by C. albicans infection indicated an abnormal intestinal homoeostasis. Similarly, C. albicans also induced the reduction of alpha diversity in fungi and caused the decrease of some other fungi levels, which indicated that C. albicans overgrowth might inhibit the growth of some fungi in the gut.
Microbiota is reported to affect the development and homoeostasis of γδT cells, and vice versa, γδT cells play important roles in the selection and maintenance of commensal microbiota (Papotto et al. Citation2021). Our study demonstrated that the in vivo γδT cell neutralisation partly rescued the bacterial dysbiosis caused by C. albicans infection, which indicated that γδT cells might be potential henchmen for C. albicans inducing gut bacterial disorders. However, the neutralisation of γδT cells further boosted the overgrowth of Ascomycota in the gut. It is worth mentioning that microbiota-related IL-17A producing γδT cells were reported to promote OC and colorectal cancers (Reis et al. Citation2022; Wei et al. Citation2022). That’s to say, γδT cells may be important parts in maintaining microbial homoeostasis, the abnormality of which may promote cancer development.
IL-17A, an important cytokine produced at low levels in response to the resident microbiota, contributes to maintaining a healthy bacterial and fungal population on mucosa and skin (Majumder and Mcgeachy Citation2021). However, the unrestrained IL-17 signal is proposed to be associated with cancer progression (Mcgeachy et al. Citation2019). Though IL-17A is important in fighting against C. albicans (Douzandeh-Mobarrez and Kariminik Citation2019), the role of IL-17A in regulating gut microbiota during C. albicans infection is unclear. Interestingly, our results showed that the neutralisation of IL-17A during C. albicans infection further reduced the F/B ratio and boosted the overgrowth of Candida in the gut of mice with oral cancer. Thus, it is indicated that IL-17A is an important cytokine in inhibiting Candida overgrowth and maintaining intestinal homoeostasis.
C. albicans infection influenced the abundance of some bacterial species, which are closely associated with intestinal diseases and prognosis. It was shown that C. albicans infection caused the decrease of Alistipes, Clostridia UCG-014, Akkermansia, and Lachnospiraceae NK4A136 group. Correlation analysis further supported the negative relationships between C. albicans and Akkermansia. Among them, Alistipes is a controversial genus of bacteria possessing protective effects against colitis or pathogenic effects on colorectal cancer (Parker et al. Citation2020). Clostridia UCG-014 is reported to be decreased in patients with Crohn’s disease and may be associated with gut barrier function (Leibovitzh et al. Citation2022). Akkermansia muciniphila is accepted to be a promising probiotic and has important roles in regulating host functions that are disrupted in various diseases including ulcerative colitis (Derrien et al. Citation2017). Lachnospiraceae NK4A136 group is also a short chain fatty acids (SCFA)-producing bacterium, which enhances gut barrier function (Ma et al. Citation2020). Thus, the decrease of potential beneficial bacteria caused by C. albicans overgrowth may further affect the host’s immune system and intestinal mucosal barrier.
Accumulating evidence has demonstrated the importance of gut microbiota in the prognosis of various types of cancers (Helmink et al. Citation2019; Sepich-Poore et al. Citation2021). Whether C. albicans influences the efficacy of tumour treatment is rarely reported. From our results, it is speculated that C. albicans infection may influence the efficacy of tumour treatment by disrupting intestinal homoeostasis. Akkermansia muciniphila was reported to be associated with the clinical benefit of PD-1 blockade in patients with non-small-cell lung cancer or kidney cancer (Derosa et al. Citation2022). Additionally, Bifidobacterium spp. and Faecalibacterium spp. were demonstrated to be associated with favourable anticancer immune responses (Routy et al. Citation2018a). Thus, the reduced abundance of Akkermansia, Bifidobacterium, and Faecalibacterium during C. albicans overgrowth may indicate an impaired anticancer immune efficacy.
Though many studies have demonstrated the role of bacteria in cancer treatment, relatively few studies explored fungi. Some recent studies provided evidence supporting fungal roles in cancer treatment. It was reported that the depletion of fungi enhanced the responsiveness of breast cancer and melanoma to radiotherapy (Shiao et al. Citation2021). Additionally, the enriched presence of Filobasidium spp. in donor faeces was demonstrated to be associated with the beneficial response to faecal microbiota transplantation (FMT) for patients with ulcerative colitis (van Thiel et al. Citation2022), which fungus was reduced during C. albicans overgrowth in our study. Thus, it would make sense to further explore the role of fungi including C. albicans in cancer treatment.
The limitations of this study should be noticed. Firstly, this study only focused on the roles of γδT cells and IL-17A in C. albicans influencing gut microbiota, but did not explore the other immune factors, such as macrophages, Th17 cells, and B cells. Secondly, the interplay between fungi and bacteria in the host is complex, further studies should explore this matter from multi-dimensional time and space. Lastly, different levels and statuses of C. albicans infection may contribute differently to intestinal immunity and intestinal flora, which should be considered.
5. Conclusions
In conclusion, this study presented the disturbed intestinal flora caused by C. albicans infection in the context of oral cancer. C. albicans overgrowth inhibited the growth of some beneficial bacteria, such as Lachnospiraceae NK4A136 group and Akkermansia, which indicated that C. albicans overgrowth during cancer development and treatment might damage the intestinal mucosal barrier and even impair the efficacy of cancer treatment. Additionally, it was speculated that immune components such as γδT cells and IL-17A might participate in the maintenance of intestinal homoeostasis. Thus, it is suggested to control C. albicans infection during the period of cancer treatment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Akuzum B, Lee JY. 2022. Context-dependent regulation of type 17 immunity by microbiota at the intestinal barrier. Immune Netw. 22(6):e46. doi: 10.4110/in.2022.22.e46.
- Aly AM, Adel A, El-Gendy AO, Essam TM, Aziz RK. 2016. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 8(1):42. doi: 10.1186/s13099-016-0124-2.
- An J, Kwon H, Kim YJ. 2023. The Firmicutes/Bacteroidetes ratio as a risk factor of breast cancer. J Clin Med. 12(6):2216. doi: 10.3390/jcm12062216.
- Bertolini M, Ranjan A, Thompson A, Diaz PI, Sobue T, Maas K, Dongari-Bagtzoglou A. 2019. Candida albicans induces mucosal bacterial dysbiosis that promotes invasive infection. PLoS Pathog. 15(4):e1007717. doi: 10.1371/journal.ppat.1007717.
- Derosa L, Routy B, Thomas AM, Iebba V, Zalcman G, Friard S, Mazieres J, Audigier-Valette C, Moro-Sibilot D, Goldwasser F, et al. 2022. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med. 28(2):315–324. doi: 10.1038/s41591-021-01655-5.
- Derrien M, Belzer C, de Vos WM. 2017. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 106:171–181. doi:10.1016/j.micpath.2016.02.005.
- Douzandeh-Mobarrez B, Kariminik A. 2019. Gut microbiota and IL-17A: Physiological and pathological responses. Probiotics Antimicrob Proteins. 11(1):1–10. doi: 10.1007/s12602-017-9329-z.
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27(16):2194–2200. doi: 10.1093/bioinformatics/btr381.
- Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. 2018. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 33(4):570–580. doi: 10.1016/j.ccell.2018.03.015.
- Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. 2019. The microbiome, cancer, and cancer therapy. Nat Med. 25(3):377–388. doi: 10.1038/s41591-019-0377-7.
- Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486(7402):207–214. doi: 10.1038/nature11234.
- Ivanov II, Tuganbaev T, AN S, Honda K. 2022. T cell responses to the microbiota. Annu Rev Immunol. 40(1):559–587. doi: 10.1146/annurev-immunol-101320-011829.
- Kumari A, Tripathi AH, Gautam P, Gahtori R, Pande A, Singh Y, Madan T, Upadhyay SK. 2021. Adhesins in the virulence of opportunistic fungal pathogens of human. Mycology. 12(4):296–324. doi: 10.1080/21501203.2021.1934176.
- Leibovitzh H, Lee SH, Xue M, Raygoza Garay JA, Hernandez-Rocha C, Madsen KL, Meddings JB, Guttman DS, Espin-Garcia O, Smith MI, et al. 2022. Altered gut microbiome composition and function are associated with gut barrier dysfunction in healthy relatives of patients with Crohn’s disease. Gastroenterology. 163(5):1364–1376. doi:10.1053/j.gastro.2022.07.004.
- Longa CMO, Nicola L, Antonielli L, Mescalchin E, Zanzotti R, Turco E, Pertot I. 2017. Soil microbiota respond to green manure in organic vineyards. J Appl Microbiol. 123(6):1547–1560. doi: 10.1111/jam.13606.
- Ma L, Ni Y, Wang Z, Tu W, Ni L, Zhuge F, Zheng A, Hu L, Zhao Y, Zheng L, et al. 2020. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes. 12(1):1–19. doi: 10.1080/19490976.2020.1832857.
- Magoc T, Salzberg SL. 2021. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27(21):2957–2963. doi: 10.1093/bioinformatics/btr507.
- Majumder S, Mcgeachy MJ. 2021. IL-17 in the pathogenesis of disease: good intentions gone awry. Annu Rev Immunol. 39:537–556. doi:10.1146/annurev-immunol-101819-092536.
- Mcgeachy MJ, Cua DJ, Gaffen SL. 2019. The IL-17 family of cytokines in health and disease. Immunity. 50(4):892–906. doi: 10.1016/j.immuni.2019.03.021.
- Mengesha BG, Conti HR. 2017. The role of IL-17 in protection against mucosal Candida infections. J Fungi (Basel). 3(4):52. doi: 10.3390/jof3040052.
- Mohamed N, Litlekalsøy J, Ahmed IA, Martinsen EMH, Furriol J, Javier-Lopez R, Elsheikh M, Gaafar NM, Morgado L, Mundra S, et al. 2021. Analysis of salivary mycobiome in a cohort of oral squamous cell carcinoma patients from Sudan identifies higher salivary carriage of Malassezia as an independent and favorable predictor of overall survival. Front Cell Infect Microbiol. 11:673465. doi: 10.3389/fcimb.2021.673465.
- Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, et al. 2017. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 5(1):153. doi: 10.1186/s40168-017-0373-4.
- Panghal M, Kaushal V, Kadayan S, Yadav JP. 2012. Incidence and risk factors for infection in oral cancer patients undergoing different treatments protocols. BMC Oral Health. 12:22. doi:10.1186/1472-6831-12-22.
- Papotto PH, Yilmaz B, Silva-Santos B. 2021. Crosstalk between gammadelta T cells and the microbiota. Nat Microbiol. 6(9):1110–1117. doi: 10.1038/s41564-021-00948-2.
- Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. 2020. The genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 11:906. doi:10.3389/fimmu.2020.00906.
- Reis BS, Darcy PW, Khan IZ, Moon CS, Kornberg AE, Schneider VS, Alvarez Y, Eleso O, Zhu C, Schernthanner M, et al. 2022. TCR-Vgammadelta usage distinguishes protumor from antitumor intestinal gammadelta T cell subsets. Sci. 377(6603):276–284. doi:10.1126/science.abj8695.
- Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. 2018a. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 15(6):382–396. doi: 10.1038/s41571-018-0006-2.
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. 2018b. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Sci. 359(6371):91–97. doi:10.1126/science.aan3706.
- Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. 2021. The microbiome and human cancer. Sci. 371(6536):eabc4552. doi: 10.1126/science.abc4552.
- Shiao SL, Kershaw KM, Limon JJ, You S, Yoon J, Ko EY, Guarnerio J, Potdar AA, McGovern DPB, Bose S, et al. 2021. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. 39(9):1202–1213. doi:10.1016/j.ccell.2021.07.002.
- Shiromizu CM, Jancic CC. 2018. Gammadelta T lymphocytes: An effector cell in autoimmunity and infection. Front Immunol. 9:2389. doi:10.3389/fimmu.2018.02389.
- Stojanov S, Berlec A, Strukelj B. 2020. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 8(11):1715. doi: 10.3390/microorganisms8111715.
- Takiishi T, Fenero C, Camara N. 2017. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. 5(4):e1373208. doi: 10.1080/21688370.2017.1373208.
- Ting NL, Lau HC, Yu J. 2022. Cancer pharmacomicrobiomics: Targeting microbiota to optimise cancer therapy outcomes. Gut. 71(7):1412–1425. doi: 10.1136/gutjnl-2021-326264.
- Umazume M, Ueta E, Osaki T. 1995. Reduced inhibition of Candida albicans adhesion by saliva from patients receiving oral cancer therapy. J Clin Microbiol. 33(2):432–439. doi: 10.1128/jcm.33.2.432-439.1995.
- Vaiserman A, Romanenko M, Piven L, Moseiko V, Lushchak O, Kryzhanovska N, Guryanov V, Koliada A. 2020. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 20(1):221. doi: 10.1186/s12866-020-01903-7.
- van Thiel IAM, Rahman S, Hakvoort TBM, Davids M, Verseijden C, van Hamersveld PHP, Bénard MV, Lodders MH, Boekhout T, van den Wijngaard RM, et al. 2022. Fecal Filobasidium is associated with clinical remission and endoscopic response following fecal microbiota transplantation in mild-to-moderate ulcerative colitis. Microorganisms. 10(4):737. doi: 10.3390/microorganisms10040737.
- Wang F, Ye Y, Xin C, Liu F, Zhao C, Xiang L, Song Z. 2021. Candida albicans triggers qualitative and temporal responses in gut bacteria. J Mycol Med. 31(3):101164. doi: 10.1016/j.mycmed.2021.101164.
- Wang X, Wu S, Wu W, Zhang W, Li L, Liu Q, Yan Z. 2023a. Candida albicans promotes oral cancer via IL-17A/IL-17RA-macrophage axis. mBio. 14(3):e0044723. doi: 10.1128/mbio.00447-23.
- Wang X, Zhang W, Wu W, Wu S, Young A, Yan Z. 2023b. Is Candida albicans a contributor to cancer? A critical review based on the current evidence. Microbiol Res. 272:127370. doi:10.1016/j.micres.2023.127370.
- Wei W, Li J, Shen X, Lyu J, Yan C, Tang B, Ma W, Xie H, Zhao L, Cheng L, et al. 2022. Oral microbiota from periodontitis promote oral squamous cell carcinoma development via gammadelta T cell activation. mSystems. 7(5):e46922. doi:10.1128/msystems.00469-22.
- Zaongo SD, Ouyang J, Isnard S, Zhou X, Harypursat V, Cui H, Routy JP, Chen Y. 2023. Candida albicans can foster gut dysbiosis and systemic inflammation during HIV infection. Gut Microbes. 15(1):2167171. doi: 10.1080/19490976.2023.2167171.
- Zhou QH, Wu FT, Pang LT, Zhang TB, Chen Z. 2020. Role of gammadelta T cells in liver diseases and its relationship with intestinal microbiota. World J Gastroenterol. 26(20):2559–2569. doi: 10.3748/wjg.v26.i20.2559.

