ABSTRACT
The order Agaricales was divided into eight suborders. However, the phylogenetic relationships among some suborders are largely unresolved, and the phylogenetic positions and delimitations of some taxa, such as Sarcomyxaceae and Tricholomopsis, remain unsettled. In this study, sequence data of 38 genomes were generated through genome skimming on an Illumina sequencing system. To anchor the systematic position of Sarcomyxaceae and Tricholomopsis, a phylogenetic analysis based on 555 single-copy orthologous genes from the aforementioned genomes and 126 publicly accessible genomes was performed. The results fully supported the clustering of Tricholomopsis with Phyllotopsis and Pleurocybella within Phyllotopsidaceae, which formed a divergent monophyletic major lineage together with Pterulaceae, Radulomycetaceae, and Macrotyphula in Agaricales. The analysis also revealed that Sarcomyxaceae formed a unique major clade. Therefore, two new suborders, Phyllotopsidineae and Sarcomyxineae, are proposed for the two major lineages. Analyses of 450 single-copy orthologous genes and four loci suggested that Tricholomopsis consisted of at least four clades. Tricholomopsis is subsequently subdivided into four distinct sections. Seventeen Tricholomopsis species in China, including six new species, are reported. Conoloma is established to accommodate T. mucronata. The substrate preference of Tricholomopsis species and the transitions of the pileate ornamentations among the species within the genus are discussed.
1. Introduction
Agaricales Underw. is one of the most diverse orders of mushroom-forming fungi (Bánki et al. Citation2022). Based on multi-locus phylogenetic analyses (Matheny et al. Citation2006; Lodge et al. Citation2014; Zhao et al. Citation2017), the positions of many taxa in Agaricales have been anchored. To date, 8 suborders, 46 families, 482 genera, and more than 40,000 species have been allocated to the order (Vizzini et al. Citation2019; Kalichman et al. Citation2020; Mou and Bau Citation2021; Bánki et al. Citation2022).
According to Dentinger et al. (Citation2016), Agaricales was phylogenetically divided into seven suborders: Agaricineae Aime, Dentinger & Gaya, Pluteineae Aime, Dentinger & Gaya, Tricholomatineae Aime et al., Marasmiineae Aime et al., Schizophyllineae Aime et al., Pleurotineae Aime et al., and Hygrophorineae Aime, et al. Subsequently, Olariaga et al. (Citation2020) further split Hygrophorineae into two suborders and additionally established Clavariineae Olariaga et al. based on six gene-loci analysis. Meanwhile, Olariaga et al. (Citation2020) also broadened the taxonomic range of Pleurotineae so that it encompassed not only the previously recognised Pleurotaceae Kühner and Pterulaceae Corner but also Phyllotopsidaceae Olariaga et al., Radulomycetaceae Leal-Dutra et al., Stephanosporaceae Oberw. & E. Horak, Sarcomyxaceae Olariaga et al., and Typhulaceae Jülich. However, the monophyletic clade Pleurotineae was less or not supported by Bayesian posterior probabilities (PP = 0.95) or maximum likelihood bootstrap proportion (ML-BP < 70%) (Olariaga et al. Citation2020), owing to the insufficient gene fragments.
Sarcomyxa P. Karst. includes four saprophytic and fan- or kidney-shaped species with prominent cheilocystidia and amyloid basidiospores (Cai et al. Citation2023). The genus was regarded as a new family based on a single species and was added to the suborder Pleurotineae (Olariaga et al. Citation2020).
The genus Tricholomopsis Singer has an undetermined phylogenetic affinity in Agaricales. Tricholomopsis includes a group of saprophytic, tricholomoid fungi with prominent cheilocystidia and is usually associated with decaying wood, growing on conifer wood, hardwood, or bamboo. Several studies have attempted to identify the phylogenetic position of the genus. Singer (Citation1939) placed Tricholomopsis in Tricholomataceae R. Heim ex Pouzar based on its morphology. However, Moncalvo et al. (Citation2002) clustered Tricholomopsis rutilans (Schaeff.) Singer and T. aurea (Beeli) Desjardin & B.A. Perry with Clavulinopsis fusiformis (Sowerby) Corner, whereas Matheny et al. (Citation2006) clustered Tricholomopsis decora (Fr.) Singer with Amanita Pers. (Lodge et al. Citation2014) suggested that T. rutilans and T. decora were close to Typhula (Pers.) Fr., Macrotyphula R.H. Petersen, and Phyllotopsis E.-J. Gilbert & Donk ex Singer, and located in the basal clade to the hygrophoroid group. Kalichman et al. (Citation2020) placed Tricholomopsis in Typhulaceae in the suborder Hygrophorineae along with Typhula, Macrotyphula, Pleurocybella Singer, and Phyllotopsis, whereas Olariaga et al. (Citation2020) established the family Phyllotopsidaceae to accommodate Phyllotopsis, Pleurocybella, and Macrotyphula in the suborder Pleurotineae without discussing Tricholomopsis. These previous studies indicate that Tricholomopsis might be related to Tricholomataceae, Clavulinopsis, Amanita, Typhulaceae, or Phyllotopsidaceae. However, previous work has not provided firm evidence for the phylogenetic position of Tricholomopsis.
Tricholomopsis was once divided into two sections according to the colour of the pileus and lamellae: sect. Tricholomopsis and sect. Platyphyllae Singer (Singer Citation1943). Later, Smith (Citation1960) further expanded the classification of Tricholomopsis by adding two additional sections based on the pileus colour: sect. Lividae A. H. Smith and sect. Decoramentum A. H. Smith. Sect. Platyphyllae was recently reassigned to Megacollybia Kotl. & Pouzar (Hughes et al. Citation2007). Jayawardena et al. (Citation2022) divided Tricholomopsis into five clades: Rubroaurantiaca, Decora, Rutilans, Scabra, and Aurea. To date, approximately 40 species have been recognised from Africa (Pegler Citation1977; Desjardin Citation2017), Asia (Hongo Citation1959, Citation1960, Citation1966; He Citation1989; Liu Citation1994; Hosen et al. Citation2020; Mao et al. Citation2021; Wang and Yang Citation2023), Australia (Horak Citation1971; Cooper and Park Citation2016), Europe (Singer Citation1939; Holec Citation2009; Olariaga et al. Citation2015; Vizzini et al. Citation2019), North America (Singer Citation1943, Citation1951; Thiers Citation1958; Smith Citation1960), and South America (Singer Citation1953, Citation1989; Horak Citation1980; Pegler Citation1983; Garrido Citation1988; Singer et al. Citation1990; Jayawardena et al. Citation2022). In China, 14 species of Tricholomopsis, including eight novel species (T. shulanensis X. He, T. lividipileata P.G. Liu, T. nigrosquamosa P.G. Liu, T. rubroaurantiaca Hosen & T.H. Li, T. galeata L. Fan & N. Mao, T. pallidolutea L. Fan & N. Mao, T. mitirubicunda L. Fan & N. Mao, and T. mucronata Zhu L. Yang & G.S. Wang) and one novel variety (T. bambusina var. megaspora P.G. Liu) have been reported (He Citation1989; Liu Citation1994; Wang et al. Citation2018; Hosen et al. Citation2020; Mao et al. Citation2021; Wang and Yang Citation2023). However, species diversity is underestimated because of limited sampling and the morphological similarity of the different taxa.
This study aimed (1) to construct a phylogenetic framework of Agaricales using a robust phylogeny; (2) to clarify the phylogenetic positions of Sarcomyxaceae and Tricholomopsis; and (3) to elucidate the infrageneric relationships and species diversity within Tricholomopsis, and to infer the evolution of substrate preference and the pileate ornamentations of the species within Tricholomopsis.
2. Materials & methods
2.1. Taxon sampling
Ninety-eight samples collected during the last two decades, including 92 Tricholomopsis, three Phyllotopsis, one Macrotyphula, and two Sarcomyxa samples, were included in this study. Voucher specimens were deposited in the Cryptogamic Herbarium of the Herbaria of Kunming Institute of Botany, Chinese Academy of Sciences (KUN-HKAS), the Mycological Herbarium of Hunan Normal University (MHHNU), and the Herbarium of Mycological Institute of Jilin Agricultural University (HMJAU). Detailed information is provided in Supplementary Table S1.
2.2. Morphological observations
Macroscopic descriptions were based on field notes and digital images of basidiomata. Colors used in the descriptions are based on standard plates (Kornerup and Wanscher Citation1978). Microscopic characteristics were observed using the hand-sectioned tissues immersed in 5% KOH and stained in Congo red solution when necessary. The size of basidiospores is presented as (a–) b – c (–d). The range b – c contains a minimum of 90% of the measured values, whereas a and d refer to the extremities of all measurements. Q indicates the length/width ratio of basidiospores in the side view, with Qm representing the average Q of all basidiospores ± standard deviation. Descriptive terms are based on the work of Vellinga and Noordeloos (Citation2001). The generic names used in this study are abbreviated as follows: “T.” for Tricholomopsis, “Ta.” for Tricholoma, “C.” for Conoloma, “Ph.” for Phyllotopsis, and “Pl.” for Pleurocybella.
2.3. DNA extraction, sequencing, and data processing
Sanger sequencing and next-generation sequencing were performed in this study. Protocols for DNA extraction, PCR amplification, and sequencing followed those described by Cai et al. (Citation2014) and the references therein. The following four pairs of primers were used for PCR amplification and Sanger sequencing: ITS1F and ITS4 for the internal transcribed spacer (ITS), LROR and LR5 for the nuclear ribosomal large subunit (nrLSU), EF1-983F and EF1-1567R for the translation elongation factor 1-α (tef1-α) and Am-6F and Am-7R for the DNA-directed RNA polymerase II second largest subunit 2 (rpb2). Sequencing traces were carefully verified using SeqMan NGen Version 17.2 (DNASTAR, Madison, WI, USA), and after the manual editing of the ends of the sequence, the paired-end sequences were assembled using DNASTAR. In total, 276 sequences were obtained and submitted to GenBank under the accession numbers listed in Table S1.
Following the genome skimming approach on an Illumina Hi-seq platform, 38 samples representing 24 species in four genera (Phyllotopsis, Sarcomyxa, Tricholomopsis, and Macrotyphula) were sequenced. Protocols for DNA extraction, library preparation, and library pooling for genome sequencing followed those described in Zeng et al. (Citation2018). Fastp (Chen et al. Citation2018) was used to control the data quality and preprocess fastq files. Genome assembly was conducted using SPAdes (Bankevich et al. Citation2012) with automatic K selection based on read length. The completeness of the genome assembly was evaluated using Benchmarking Universal Single-Copy Orthologs (BUSCO v3) (Manni et al. Citation2021), with the Agaricales OrthoDB v10 database (Zdobnov et al. Citation2021) as a reference. Thereafter, single-copy orthologous genes for each sample were retrieved from the genome using the results from the BUSCO analysis.
2.4. Sequence data matrix construction
Three data matrices were generated for different purposes. The first matrix (Matrix I) comprised single-copy ortholog genes from 141 genomes of representatives of Agaricales and outgroups, including 126 publicly available genomes downloaded from A-WINGS (Yamamoto et al. Citation2014), the Joint Genome Institute (JGI), and the National Center for Biotechnology Information (NCBI), and 15 genomes generated in the present study. These were used to establish the framework of suborders and the phylogenetic positions of Sarcomyxaceae and Tricholomopsis within Agaricales. The second matrix (Matrix II) included single-copy orthologous genes from 37 genomes of Tricholomopsis and three genomes of Phyllotopsis as an outgroup and was used to elucidate the infrageneric classification of Tricholomopsis species. The last matrix (Matrix III) included four-locus sequences from Tricholomopsis, Phyllotopsis, and Pleurocybella and was used to demonstrate the diversity of Tricholomopsis.
For Matrix I, we only retained single-copy orthologous genes that could be detected in 80% of the total samples and encoded proteins > 100 amino acids in length. For Matrix II, we retained genes that could be detected in 100% of samples and encoded proteins > 100 amino acids in length. Each single gene dataset was aligned using MAFFT v7.487 (Katoh and Standley Citation2013) under the L-INS-I setting, and ambiguously aligned regions were trimmed using Gblocks 0.91b (Castresana Citation2000), with the minimum length of a block set as 2, allowing the gap positions when they occurred in half of the sequences. Subsequently, all datasets were concatenated using PhyKIT (Steenwyk et al. Citation2021) to form the final matrix.
For Matrix III, each single locus dataset was automatically aligned using MAFFT v7.487 (Katoh and Standley Citation2013), manually adjusted and trimmed using Mega 7.0 (Kumar et al. Citation2016), and then concatenated using SequenceMatrix v1.8 (Vaidya et al. Citation2011). The dataset finally comprised 124 ITS sequences (761 bp), 116 nrLSU sequences (942 bp), 42 rpb2 sequences (821 bp), and 79 tef1-α sequences (615 bp) from 155 representative samples (Table S1).
2.5. Phylogenetic analysis
For Matrix I, inconsistencies between genes (Shen et al. Citation2021) were first examined before performing the phylogenetic analysis. First, a single concatenated tree was constructed with the concatenated data matrix using IQ-TREE 2 (Minh et al. Citation2020) based on the single model LG+G4. For each gene, a gene tree was inferred using IQ-TREE 2 with automatic detection for the best-fitting model with the “−MFP” option, and branch support was evaluated based on 1,000 replicates of the ultrafast bootstrap. Low-supported branches (BS < 30%) in each gene tree were collapsed using Newick Utilities v1.6 (Junier and Zdobnov Citation2010). Subsequently, all gene trees were used to infer a coalescent tree using Astral 5.7.8 (Mirarab et al. Citation2014). Thereafter, IQ-TREE 2 and Astral 5.7.8 were used to infer the difference in gene-wise loglikelihood score (ΔGLS) and gene-wise quartet score (ΔGQS) (Shen et al. Citation2021) based on concatenated and coalescent trees. Genes were considered consistent when their ΔGLS and ΔGQS were both > 0 or < 0. Finally, 555 consistent genes were filtered for the next analysis (Table S3). The final tree was built using IQ-TREE 2 with the LG+G4 model. BP (Kishino et al. Citation1990), one-sided Kishino-Hasegawa (Kishino and Hasegawa Citation1989), Shinodaira-Hasegawa (Shimodaira and Hasegawa Citation1999), expected likelihood weight (Strimmer and Rambaut Citation2002) and approximately unbiased (Shimodaira and Goldman Citation2002) topology tests were conducted using IQ-TREE 2 (Minh et al. Citation2020) with 10,000 RELL replicates. For Matrix II, we followed the analysis process of Matrix I, and 450 genes were filtered for the final tree.
For Matrix III, ML and Bayesian inference (BI) algorithms were employed. MrModeltest 2.3 (Nylander Citation2004) was used to identify the optimal model for each dataset based on the Akaike information criterion. The ML analysis was performed using raxmlGUI 2.0 beta (Edler et al. Citation2021) with the default parameters and bootstrapping with 1,000 replicates. The BI analysis was performed using MrBayes v3.2 (Ronquist et al. Citation2012) with the Markov chain Monte Carlo method. Three independent runs with four chains each were run for 3 million generations, and trees were sampled every 100 generations. Chain convergence was determined using Tracer v1.7 (Rambaut et al. Citation2018) to ensure sufficiently large effective sample sizes (>200). Trees were summarised and statistical values were obtained using the sump and sumt commands in MrBayes by discarding the first 25% of generations as burn-ins. The genealogical concordance phylogenetic species recognition (GCPSR) criterion (Taylor et al. Citation2000) was employed to infer species boundaries.
The ancestral states of pileate ornamentations and substrate preference were constructed using ape (Paradis et al. Citation2019) and phytools (Revell Citation2012) packages in R with stochastic character mapping 1,000 times under an equal-rates model based on the phylogenetic tree of Matrix II.
2.6. Data and code availability
The genomes of Tricholomopsis generated in this study were submitted to the NCBI database. Other genome assemblies were downloaded from the NCBI or JGI (Table S2). All scripts, data matrices, and phylogenetic trees were deposited on GitHub (https://github.com/jungleblack007/Tricholomopsis).
3. Results
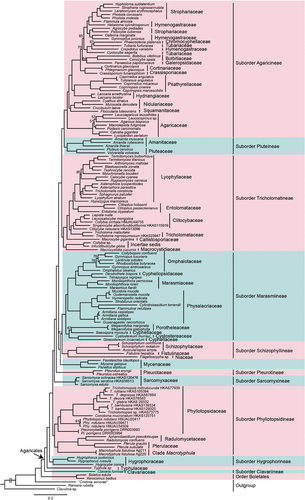
The ML tree of Agaricales inferred from 555 single-copy orthologous genes is shown in . As indicated in the tree, Agaricales is monophyletic in origin with 100% BS support. This order is further divided into 12 well-supported major clades, including eight previously recognised suborders (Agaricineae, Pluteineae, Tricholomatineae, Marasmiineae, Schizophyllineae, Pleurotineae s. str., Hygrophorineae, and Clavariineae), a clade composing Mycenaceae, and three suborder-level lineages separated from suborder Pleurotineae s. l. The first lineage, representing the family Typhulaceae, is located between the suborders Hygrophorineae and Clavariineae. The second lineage comprises the families Phyllotopsidaceae, Radulomycetaceae, and Pterulaceae and genus Macrotyphula, which diverged from Hygrophoraceae. The third lineage represents the family Sarcomyxaceae. Interestingly, Tricholomopsis, along with Phyllotopsis and Pleurocybella, are within the family Phyllotopsidaceae with full statistical support.
Figure 1. Phylogeny of Agaricales inferred from a matrix containing 555 single-copy orthologs based on the maximum likelihood (ML) analysis method. Nodes without numeric labels are supported with 100% BS.
To investigate whether the suborder Pleurotineae, second lineage, and third lineage were three truly separate major clades, 15 alternative topologies for the relationship between the three clades and other clades within Agaricales were evaluated (). Only topology 1 was accepted, while the other 14 topologies were rejected by all the topology tests (), indicating that the three clades are certainly independent.
Figure 2. Fifteen alternative topologies of Agaricales. Out, outgroup and basal clades of Agaricales (including Hygrophorineae, Typhulaceae, Clavariineae, and outgroups); Phy, Phyllotopsidineae; Sar, Sarcomyxineae; Ple, Pleurotineae; Oth, other clades in Agaricales (including Agaricineae, Pluteineae, Tricholomatineae, Marasmiineae, and Schizophyllineae).
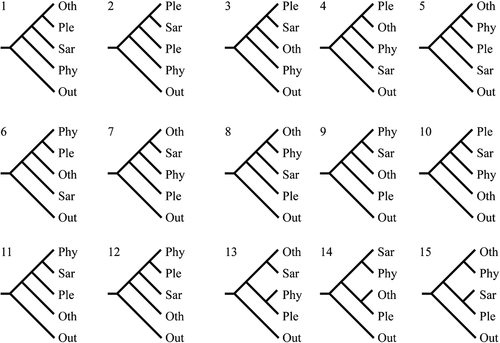
Table 1. Topology tests of 15 alternative hypotheses for the relationships among Sarcomyxineae, Pleurotineae, Phyllotopsidineae, and the other clades within Agaricales.
The ML tree of Tricholomopsis based on 450 single-copy orthologous genes is shown in . Tricholomopsis s. l. is divided into two deeply divergent clades. One represents the genus Tricholomopsis s. str., and the other forms a genus-level clade corresponding to the novel genus Conoloma. Tricholomopsis s. str. is divided into four clades, which are recognised as sections in this study.
Figure 3. (a) phylogeny of Tricholomopsis inferred from a matrix containing 450 single-copy orthologous genes using the maximum likelihood (ML) analysis method. Nodes without numeric labels are supported with 100 BS. (b) phylogeny of Tricholomopsis inferred from a matrix consisting of ITS-nrLSU-rpb2-tef1-α using Bayesian inference (BI) analysis. BI (BPP ≥0.90) and ML bootstrap support values (ML ≥ 70) are shown (BPP/ML). The habitats and ornamentations of pileus are labelled after each species.
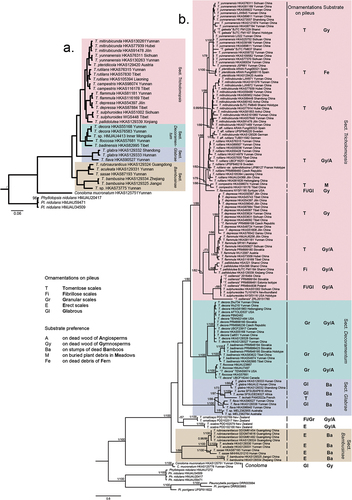
Both ML and BI phylogenetic analyses based on four gene loci produced similar tree topologies; therefore, only the tree inferred from the Bayesian analysis is shown in . As indicated in the tree, Tricholomopsis is divided into five clades, four of which correspond to the aforementioned sections of Tricholomopsis, and the remaining includes T. ornaticeps and T. scabra from New Zealand, which were not included in the analysis shown in because of the lack of genomic data. Based on the grouping and ranking criteria of the GCPSR method, 23 species of Tricholomopsis were delimited, and one single species of Conoloma was recognised.
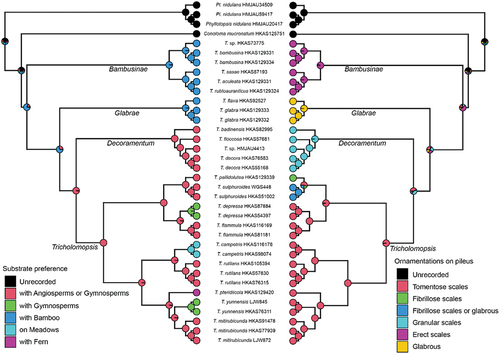
Ancestral state reconstruction of Tricholomopsis () inferred that these species preferred Bamboo stumps. Subsequently, a transition occurred towards a preference for rotten wood or buried debris in forests of either Angiosperms or Gymnosperms. Finally, in the recently diverged lineages, some species gradually evolved a preference for growing on ferns or in meadows. The ancestral ornamentation on the pileus of Tricholomopsis was characterised by erect scales. Diversification occurred over time, resulting in various types of ornamentation, including glabrous, fibrillose, and granular scales. Ultimately, a derived state with tomentose scales became fixed in the recently diverged lineages.
Figure 4. Ancestral state reconstructions of substrate preference (left part) and ornamentations on pileus (right part) of Tricholomopsis using ape and phytools packages in R. The phylogenetic tree was based on the results of 450 single-copy orthologous genes, and pie chart of each node were summarised by stochastic character mapping under equal-rates model with 1,000 times. Names for each section in Tricholomopsis are labelled next to the corresponding branches.
4. Discussion
4.1. Phylogenetic framework of Agaricales and relationships among suborders
We generated the most comprehensive and robust phylogenetic framework for Agaricales to date through an in-depth phylogenomic analysis including 42 families and two family-level lineages (). Ten suborders and one suborder-level lineage could be recognised in Agaricales, which were almost the same as those reported in previous studies on recently diverged lineages (Dentinger et al. Citation2016; Olariaga et al. Citation2020). However, Mycenaceae Overeem, once arranged in Marasmiineae with less support (ML-BP < 70), forms a clade sister to Marasmiineae and Schizophyllineae. The suborder Pleurotineae initially included only Pleurotaceae and Pterulaceae (Dentinger et al. Citation2016), but was expanded by Olariaga et al. (Citation2020) to encompass five additional families: Phyllotopsidaceae, Radulomycetaceae, Stephanosporaceae, Sarcomyxaceae, and Typhulaceae. In our study, Pleurotineae is delimited to include only Pleurotaceae, whereas Phyllotopsidaceae, Radulomycetaceae, and Pterulaceae are grouped as a separate suborder (see Taxonomy section). Sarcomyxaceae, typified by Sarcomyxa serotina (Pers.) V. Papp, is considered an independent suborder here due to its unique morphological features and phylogenetic position (see Taxonomy section). Typhulaceae, typified by Typhula incarnata Lasch (Olariaga et al. Citation2022), was once also arranged in Pleurotineae s. l., but is identified as an independent suborder-level lineage in this study. However, the sampling of the family was not comprehensive enough to regard it as a new suborder (Shen et al. Citation2023). Additional studies are required to improve our understanding of the phylogenetic positions of the family Stephanosporaceae and the genus Macrotyphula.
Dentinger et al. (Citation2016) treated Hygrophoraceae and Clavariaceae within the suborder Hygrophorineae. However, the findings of Olariaga et al. (Citation2022) and the present study revealed that these two families do not represent a monophyletic group but rather two independent lineages, namely the suborders Hygrophorineae and Clavariineae. A comparison of the suborders in Agaricales among the three treatments (Dentinger et al. Citation2016; Olariaga et al. Citation2020; this study) is shown in .
Table 2. Comparisons of suborder divisions within Agaricales by different authors.
4.2. Phylogenetic position of Tricholomopsis and delimitation of Phyllotopsidaceae
In this study, we confirmed that Tricholomopsis is clustered within the family Phyllotopsidaceae of the suborder Phyllotopsidineae (see Taxonomy section for Phyllotopsidineae). Tricholomopsis mucronata Zhu L. Yang & G.S. Wang was distinct from the other Tricholomopsis species, indicating that it represents a separate genus (). Morphologically, it differs from Tricholomopsis species by its mucronate umbo of the pileus and a cortiniform annulus on the upper part of the stipe. Therefore, the new genus, Conoloma (see Taxonomy section) is proposed to accommodate this unique species. The genus Macrotyphula, which was arranged in the same family with less support (Olariaga et al. Citation2020), forms a family-level sister clade to Pterulaceae and Radulomycetaceae. Therefore, the genus might be distinguished from Phyllotopsidaceae, but understanding whether it should be arranged as a separate family requires more in-depth analysis with more samples. Finally, Phyllotopsidaceae comprises Conoloma, Phyllotopsis, Pleurocybella, and Tricholomopsis. The common features of this family are the fleshy basidioma; hyaline, smooth, and non-amyloid basidiospores; prominent cheilocystidia; and the presence of clamp connections in the basidioma.
4.3. Delimitation and evolution of infrageneric sections in Tricholomopsis
Molecular phylogenetic analyses based on the 450 single-copy orthologous genes concatenated dataset () and ITS-nrLSU-tef1-α-rpb2 concatenated dataset () supported four sections in Tricholomopsis, including sect. Tricholomopsis, sect. Decoramentum, sect. Glabrae, and sect. Bambusinae (see Taxonomy part). Among the four identified sections, sect. Tricholomopsis and sect. Decoramentum were mostly congruent with the work of Smith (Citation1960), while T. sulphureoides and T. flavescens were moved from sect. Tricholomopsis to sect. Decoramentum. Sect. Glabrae and sect. Bambusinae are newly proposed in this study. includes the comparisons among the four infrageneric treatments of Tricholomopsis (Singer Citation1943; Smith Citation1960; Jayawardena et al. Citation2022; this study).
Table 3. Section divisions within Tricholomopsis by different authors.
Owing to the lack of genomic data, the phylogenomic four-section framework () did not include two New Zealand species, T. scabra and T. ornaticeps (Cooper and Park Citation2016), which might belong to a unique section-level clade (), or the sect. Lividae proposed by Smith (Citation1960). However, sect. Lividae can be easily distinguished from the above-mentioned sections by the livid colour of the pileus and lamellae and larger pleurocystidia. The phylogenetic positions of T. scabra and T. ornaticeps and T. sect. Lividae require further investigation.
Our data also revealed that two unique characteristics, substrate preference and ornamentations on the pileus surface, could help distinguish the four sections within Tricholomopsis. These distinctive features serve as valuable taxonomic markers, aiding in the classification and differentiation of various sections within the genus. Moreover, the transition of substrate preference from bamboo to Angiosperm/Gymnosperm forests as well as grasslands and Pteridium aquilinum may provide a plausible explanation for the radiation and ecological niche differentiation observed in Tricholomopsis to some extent (Sato et al. Citation2017; Han et al. Citation2020; Li et al. Citation2020). However, to our knowledge, no previous studies have provided evidence of the ornamentation of pileus contributing to fungal adaptation to the environment. Nevertheless, the pileate ornamentation of Tricholomopsis, transitioning from erect scales to glabrous or fibrillose/granular scales and ultimately to tomentose scales, undoubtedly holds some degree of significance for the adaptation of fungi to their surroundings. These intriguing features of Tricholomopsis highlight the multifaceted nature of fungal adaptation and evolution. Further investigations into the genetic, ecological, and functional aspects of these characteristics may provide deeper insights into the ecological success and diversification of Tricholomopsis species.
5. Taxonomy
Phyllotopsidineae Zhu L. Yang & G. S. Wang, subord. nov.
MycoBank: MB 849,776.
Type genus: Phyllotopsis E.-J. Gilbert & Donk ex Singer Citation1936.
Etymology: From the type genus Phyllotopsis.
Diagnosis: Phyllotopsidineae differs from other suborders of Agaricales by its saprophytic nutrient mode; usually small non-amyloid basidiospores; and the common presence of clamp connections and cheilocystidia.
Description: Basidioma corticoid, tricholomoid, pleurotoid, or clavarioid and sometimes arising from a sclerotium, small to large, fleshy or tough. Basidia cylindrical to clavate, usually 4-spored. Basidiospores small, globose to ellipsoid, cylindrical or allantoid, colourless and hyaline, smooth, and non-amyloid. Cheilocystidia usually present in tricholomoid and pleurotoid genera, and pleurocystidia sometimes present in tricholomoid genera. Pileipellis a cutis with a transition to a trichoderm in tricholomoid and pleurotoid genera. Clamp connections usually present.
Known distribution: Probably nearly cosmopolitan.
Habitat: Saprotrophic, on rotten wood above the ground or buried substrates.
Families included: Phyllotopsidaceae Olariaga et al. (Citation2020), Pterulaceae Corner (1971), and Radulomycetaceae Leal-Dutra et al. (Citation2020).
Notes: In previous work, these three families and the genus Macrotyphula were once treated in the suborder Pleurotineae (Dentinger et al. Citation2016; Kalichman et al. Citation2020; Olariaga et al. Citation2020). Based on the phylogenomic analysis in this study, these four taxa formed a lineage separated from the suborder Pleurotineae. Thus, it is considered as a new suborder.
Phyllotopsidaceae Olariaga et al., Stud. Mycol. 96: 175 (2020)
Type genus: Phyllotopsis Singer Citation1936.
Description: Basidioma pleurotoid or tricholomoid, small to large, flesh. Spore prints white or pale pink. Basidia cylindrical to clavate, usually 4-spored. Basidiospores small, globose to ellipsoid, cylindrical, or allantoid, colourless and hyaline, smooth, and non-amyloid. Cheilocystidia usually present and pleurocystidia sometimes present in tricholomoid genera. Pileipellis a cutis with the transition to a trichoderm. Clamp connections present.
Known distribution: Probably nearly cosmopolitan.
Habitat: Solitary or scattered on rotten wood above ground or buried substrates.
Genera included: Pleurocybella, Phyllotopsis, Conoloma, and Tricholomopsis.
Notes: Olariaga et al. (Citation2020) placed Phyllotopsis, Pleurocybella, and Macrotyphula in the family Phyllotopsidaceae. Our analyses suggested that Macrotyphula forms a separate clade. Tricholomopsis and Conoloma in the family Phyllotopsidaceae were confirmed for the first time. All four genera of the family Phyllotopsidaceae share species with a saprotrophic nutrition mode, the presence of clamp connections, and a cutis pileipellis with a transition to a trichoderm.
Conoloma Zhu L. Yang & G. S. Wang, gen. nov.
Mycobank: MB 849,787.
Type species: Conoloma mucronatum (Zhu L. Yang & G. S. Wang) Zhu L. Yang & G. S. Wang.
Etymology: The name “Conoloma” referring to the mucronate pileus of the basidioma.
Diagnosis: Conoloma differs from Tricholomopsis by its mucronate umbo of the pileus, fibrillose annuliform zone on the upper part of the stipe, and smaller cheilocystidia.
Description: Basidioma tricholomoid, small to medium-sized, fleshy. Basidia cylindrical to clavate, usually 4-spored. Basidiospores small, globose to ellipsoid, colourless and hyaline, smooth, and non-amyloid. Cheilocystidia present. Pileipellis a cutis. Clamp connections present.
Known distribution: Only known from southwestern China.
Habitat: Saprotrophic, on rotten wood above the ground or buried wood.
Notes: Conoloma mucronatum was previously recognised as a species of Tricholomopsis (Wang and Yang Citation2023). However, based on our genomic analysis, this species formed a lineage sister but is distinct from Tricholomopsis. It is therefore considered as a separate genus.
Conoloma mucronatum (Zhu L. Yang & G. S. Wang) Zhu L. Yang & G. S. Wang, comb. nov.
Mycobank: MB 849,788.
Basionym: Tricholomopsis mucronata Zhu L. Yang & G. S. Wang, in Wang & Yang, Journal of Fungal Research 21: 27 (2023).
Tricholomopsis Singer, Schweiz. Z. Pilzk. 17: 56 (1939)
Type species: T. rutilans (Schaeff.) Singer.
Diagnosis: Tricholomopsis differs from other genera of Phyllotopsidaceae by its tricholomoid basidiomata, glabrous, tomentose to fibrillose pileus, and conspicuous cheilocystidia.
Known distribution: Probably nearly cosmopolitan
Habitat: Solitary or scattered on rotten wood above ground or underground, on substrates with dead bamboo, or on buried grass or litter.
Notes: Smith (Citation1960) divided Tricholomopsis into four sections, among which T. sect. Platyphylla has been transferred to Megacollybia (Hughes et al. Citation2007), and T. sect. Lividae can be distinguished from the other sections by a livid pileus and lamellae and its large pleurocystidia. Our phylogenetic analysis confirmed the erections of sects. Tricholomopsis, Decoramentum, Bambusinae and Glabrae. There may well be another section including T. ornaticeps and T. scabra (Hosen et al. Citation2020; Jayawardena et al. Citation2022). To locate their exact position in Tricholomopsis, additional studies are necessary.
Key to the sections of Tricholomopsis in China
Occurring on dead stumps of bamboo; pleurocystidia absent or rarely present........................... 2
Occurring on other plant debris; pleurocystidia present............................................................................ 3
Pileus yellow or yellowish; scales absent or velvety.......................................................... T. sect. Glabrae
Pileus brown to dark brown; scales present and erect............................................. T. sect. Bambusinae
Pileus surface covered with purplish to red, tomentose to fibrillose scales; stipe glabrous or covered with purplish scales........................................................................... T. sect. Tricholomopsis
Pileus surface covered with greyish minute scales; stipe glabrous or covered with greyish scales...................................... T. sect. Tricholomopsis
Tricholomopsis sect. Bambusinae Zhu L. Yang & G. S. Wang, sect. nov.
Mycobank: MB 849,784.
Type: Tricholomopsis bambusina Hongo.
Diagnosis: Tricholomopsis sect. Bambusinae can be distinguished from the other sections of Tricholomopsis by its yellow, orange to brown pileus covered with erect fibrillose scales, sinuate to subdecurrent lamellae, subglobose to broadly ellipsoid basidiospores, and absence of pleurocystidia. Species of T. sect. Bambusinae prefer forests with bamboo.
Description: Basidioma medium-sized, tricholomoid; pileus applanate, densely covered with erect fibrillose scales; lamellae sinuate, adnate to subdecurrent; stipe cylindrical, hollow; basidiospores subglobose to broadly ellipsoid, smooth, inamyloid; basidia clavate, 4-spored; pleurocystidia absent; cheilocystidia prominent; pileipellis cutis to trichoderm; clamps present. Known from East Asia on stumps or buried substrate of bamboo.
Notes: Previous data and our multi-locus phylogenetic study indicated that T. bambusina, T. sasae, T. rubroaurantiaca, and T. aculeata (Hongo Citation1959, Citation1960; Hosen et al. Citation2020) formed the earliest divergent lineage of Tricholomopsis. We treated this lineage as a novel section. All species in this section live on stumps or buried substrates of bamboo.
Key to the species of Tricholomopsis sect. Bambusinae in China
Pileus dark yellow; scales dark red-brown....................................................... T. bambusina
Pileus light yellow or light orange; scales reddish-brown..................................................................... 2
Pileus light yellow; basidiospores ellipsoid to ovoid.................................................................... T. sasae
Pileus light orange; basidiospores subglobose to broadly ellipsoid..................................................... 3
Stipe > 4 cm in length; basidiospores > 6 μm in length............................................................ T. aculeata
Stipe < 4 cm in length; basidiospores < 6 μm in length............................................. T. rubroaurantiaca
Tricholomopsis aculeata Zhu L. Yang & G. S. Wang, sp. nov.
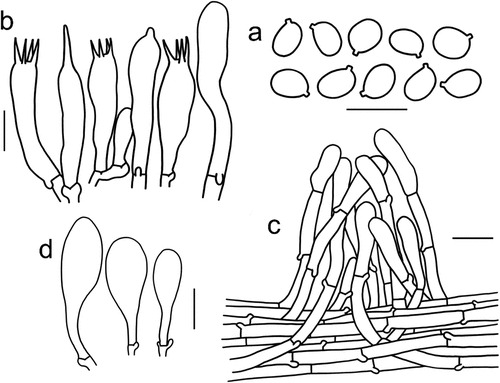
Figure 5. Microscopic features of Tricholomopsis aculeata (type, HKAS 129,330). (a) Basidiospores; (b) Hymenium; (c) Pileipellis; (d) Cheilocystidia. Bars: a – b = 10 μm, c – d = 20 μm.
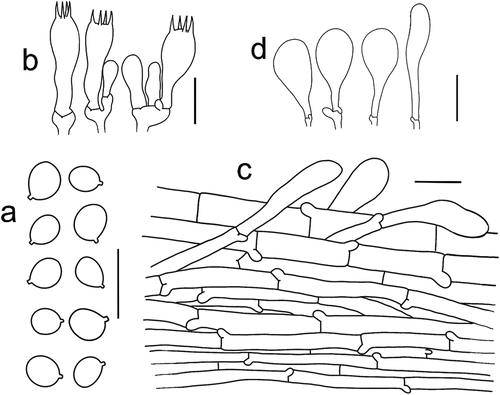
Figure 6. (a) Tricholomopsis aculeata (HKAS 129,330, photo by Gengshen Wang); (b) T. bambusina (HKAS 129,325, photo by Kuan Zhao); (c) T. sasae (HKAS 87,193, photo by Xiao-Bin Liu); (d) T. rubroaurantiaca (HKAS 129,324, photo by Jing-Wei Li). Bars: a – d = 1 cm.

Mycobank: MB 849,785.
Etymology: aculeata, referring to the erect fibrillose or scales on the pileus.
Type: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Menghai County, Bulang Mountain, 1,000 m elev., on buried debris in a forest with bamboos, 23 June 2020, Geng-Shen Wang 982 (HKAS 129,330, holotype).
Diagnosis: Tricholomopsis aculeata differs from other species of sect. Bambusinae by its yellow to orange pileus, filamentous inner veils, segmentiform to adnate lamellae, longer basidiospores (6.5–8.5 × 4–5.5 μm), and more robust basidia (22–32 × 7–10 μm).
Description: Basidiomata small to medium-sized. Pileus 2–4.5 cm in diam., at first hemispherical to convex, then applanate to plano-concave, margin straight; surface dry, brown orange to buff (5C5–8) at centre, light orange, orange-white to white (6A1–6) towards margin; surface densely covered with radially erect fibrillose or scales, brown orange (5C7–8); inner veils filamentous. Lamellae segmentiform, sinuate to adnate with decurrent teeth, crowded, edges entire, white, yellow-white, pale yellow (4A1–3) to yellow-orange (4A6–8). Stipe 5–6 × 0.4 cm, cylindrical, hollow, yellow-white, bright yellow (4A2–4) to light orange (5A3–4), glabrous.
Basidiospores [51/2/2] 6.5–8.5 × 4–5.5 μm, Q = (1.08–) 1.10–1.29 (−1.30), Qm = 1.19 ± 0.05, subglobose to broadly ellipsoid, in-amyloid, colourless, hyaline, thin-walled, smooth; apiculus relatively small. Basidia 22–32 × 7–10 μm, clavate, 4-spored, hyaline, sterigmata 2–4 μm long. Pleurocystidia absent. Cheilocystidia 32–84 × 12–24 μm, broadly cylindrical, clavate to broadly clavate, thin-walled, colourless, hyaline. Pileipellis a cutis with the transition to a trichoderm at regular intervals, composed of 4–12 µm wide, thin-walled, filamentous hyphae, terminal cells subcylindrical to clavate, 10–17 × 25–58 μm, with round apex. Clamps present at all parts of basidioma.
Known distribution: southwestern China.
Habitat: Scattered on buried substrates in forests with bamboo.
Additional material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Menghai County, Bulang Mountain, 1000 m elev., on buried debris in a forest with bamboos, 23 June 2020, Geng-Shen Wang 983 (HKAS 129,331).
Notes: Tricholomopsis aculeata is characterised by its light orange pileus with erect fibrils, while T. bambusina is a pale yellowish pileus with reddish brown scales and T. sasae is a yellowish-brown pileus with minute velvety-squamous scales. Tricholomopsis aculeata is similar to T. rubroaurantiaca but can be differentiated from the latter by its longer basidiospores and more robust basidia.
Tricholomopsis bambusina Hongo, J. Jap. Bot. 34: 239 (1959)
Known distribution: China and Japan (Hongo Citation1959; Liu Citation1994; this study).
Habitat: Scattered on buried substrates in forests with bamboo.
Material examined: China, Zhejiang Province, Lishui City, Songyang County, 122 m elev., in a forest with bamboo, 14 September 2021, Geng-Shen Wang Citation1856 (HKAS 129,334). Jiangxi Province, Jing’an County, Jiangxi Jiulingshan National Nature Reserve, 300 m elev., in a forest with bamboo, 14 July 2021, Jiulingshan 306 (HKAS 129,325).
Notes: Tricholomopsis bambusina is characterised by basidiomata covered with reddish-brown scales and small basidiospores, which differentiates it from other species in T. sect. Bambusinae.
Tricholomopsis rubroaurantiaca Hosen & T. H. Li, in Hosen, Xu, Gates & Li, Mycoscience 61(6): 343 (2020)
Known distribution: South China (Hosen et al. Citation2020).
Habitat: Solitary or in small clusters on buried substrate in forests with bamboo.
Material examined: China, Guangdong Province, Zhaoqing City, Dinghushan Nature Reserve, 76 m elev., 21 June 2016, H16062103 (HKAS 129,324).
Notes: Tricholomopsis rubroaurantiaca was described in China; for descriptions and comparisons to other species, see Hosen et al. (Citation2020).
Tricholomopsis sasae Hongo, J. Jap. Bot. 35: 85 (1960)
Known distribution: Japan and China (Hongo Citation1960; Kasuya et al. Citation2015).
Habitat: scattered or clustered on roots of bamboo-grass or buried wood in forests with Fagaceae or coniferous forests.
Material examined: China, Yunnan Province, Longlin County, Zhenan Town, 2,100 m elev., in a forest with Fagaceae, 15 March 2014, Xiao-Bin Liu 641 (HKAS 87,193). Hunan Province, Yizhang County, Mang Mountain, 1,490 m elev., on rotten wood in a coniferous forest, 30 July 2018, MHHNU 31,210.
Notes: Tricholomopsis sasae was first described in Japan by Hongo (Citation1960), and it is characterised by a yellowish-brown pileus covered with minute velvety-squamous scales. For comparisons between other species in T. sect. Bambusinae, see the notes on T. aculeata and T. bambusina.
Tricholomopsis sect. Decoramentum A. H. Smith, Brittonia 12(1): 57 (1960)
Type: Tricholomopsis decora (Fr.) Singer.
Description: Basidioma tricholomoid; pileus glabrous to covered with greyish minute fibrillose scales; basidiospores subglobose to oblong, smooth, inamyloid; cheilocystidia prominent; pileipellis cutis to trichoderm; solitary or scattered on rotten wood or soil in coniferous or broad-leaved forests. Tricholomopsis sect. Decoramentum can be distinguished from the other sections by its greyish fibrillose scales on the pileus.
Notes: Tricholomopsis sect. Decoramentum was first described by Smith (Citation1960). To date, three taxa of T. sect. Decoramentum have been recognised in the phylogenetic analyses. Our phylogenetic analysis indicated that T. sulphureoides and T. flavescens belong to sect. Tricholomopsis rather than sect. Decoramentum, as suggested by .
Key to the species of Tricholomopsis sect. Decoramentum in China
(1) Stipe is >8 mm in width; basidiospores subglobose to ellipsoid............................................................. T. floccosa
(1) Stipe is <8 mm in width; basidiospores ellipsoid........................................................................... 2
(2) Scales tiny and light; pileus dark yellow ........................................................................ T. badinensis
(2) Scales grainy and dark; pileus bright yellow................................................................... T. decora
Tricholomopsis badinensis J. Holec, M. Kolařík & V. Kunca, in Holec, Kunca & Kolařík, Mycol. Progr. 18(3): 325 (2019)
Figure 7. (a) Tricholomopsis badinensis (HKAS 83,622, photo by Qing Cai); (b) T. decora (HKAS 129,327, photo by Jianwei Liu); (c – d) T. floccosa (HKAS 57,681, photo by Gang Wu); (e) T. flava (HKAS 96,940, photo by Yan-Jia Hao 105); (f) T. glabra (HKAS 129,332, photo by Ting Guo).

Known distribution: Europe and Western China (Holec et al. Citation2019; this study).
Habitat: Scattered on rotten wood in coniferous and broad-leaved forests.
Material examined: China, Tibet Autonomous Region, Nyingchi, Bomi County, 2,462 m elev., in a forest with Fagaceae, Betulaceae, and Pinus, 2 July 2014, Yan-Jia Hao 1204 (HKAS 82,995); same locality, in a forest with Fagaceae, Betulaceae, and Pinus, 2 July 2014, Qing Cai Citation1162 (HKAS 83,622). Yunnan Province, Diqing Tibetan Autonomous Prefecture, Haba Snow Mountain, 2,800 m elev., on rotten wood, 15 August , Li-Ping Tang 643 (HKAS 54,874).
Notes: Tricholomopsis badinensis was described from Slovakia by Holec et al. (Citation2019). T. badinensis differs from T. decora by its more muted background pileus colour, lighter scales, and longer basidia, and from T. sulfureoides by minute scales on the pileus and longer and ellipsoid spores.
Tricholomopsis decora (Fr.) Singer, Schweiz. Z. Pilzk. 17: 56 (1939)
Known distribution: Europe, North America, and Asia (Smith Citation1960; Liu Citation1994; Holec and Kolařík Citation2011, Citation2013; Olariaga et al. Citation2015).
Habitat: Solitary to scattered on rotten wood in coniferous or broad-leaved forests.
Material examined: China, Heilongjiang Province, Yichun City, Yangwanghe Forest Park, 352 m elev., on rotten wood, 18 August , Xiang-Hua Wang Citation2666 (HKAS 61,863). Yunnan Province, Lijiang City, Yulong Mountain, 3,000 m elev., on rotten wood in a coniferous forest, 10 August , Qi Zhao Citation8171 (HKAS 55,168); Diqing Tibetan Autonomous Prefecture, Shangri-La, 3,611 m elev., on rotten wood, 20 January 2016, Jian-Wei Liu Citation1555 (HKAS 129,327). Austria: Kleinwalsertal, 1,300 m elev., on rotten wood in a forest with Picea and Abies, 29 September 2016, Zhu-Liang Yang Citation5962 (HKAS 129,328).
Notes: Tricholomopsis decora presents a global distribution, and is found the in Holarctic realm (Smith Citation1960; Liu Citation1994; Holec and Kolařík Citation2013; Olariaga et al. Citation2015). T. decora can be distinguished from T. floccosa by its brighter pileus, tinier scales, slender basidiomata, and more elliptical basidiospores.
Tricholomopsis floccosa Zhu L. Yang & G. S. Wang, sp. nov.
Figure 8. Microscopic features of Tricholomopsis floccosa (type, HKAS 57,681). (a) Basidiospores; (b) Hymenium; (c) Cheilocystidia; (d) Pileipellis. Bars: a – b = 10 μm, c – d = 20 μm.
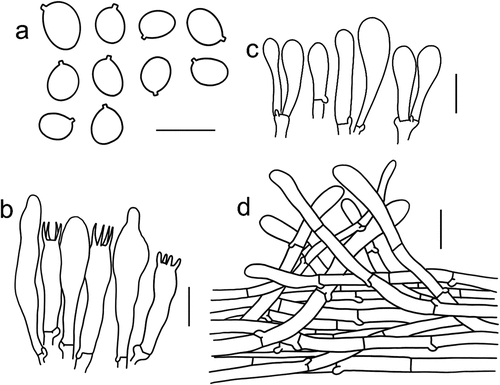
Mycobank: MB 849,779.
Etymology: floccosa referring to the floccus scales on the pileus.
Type: China, Yunnan Province, Diqing Tibetan Autonomous Prefecture, Shangri-La, 3,750 m elev., on rotten wood in a forest with Abies, 25 August, Gang Wu 149 (HKAS 57,681, holotype).
Diagnosis: Tricholomopsis floccosa differs from the other species of T. sect. Decoramentum by its tan to olive-brown scales on pileus, robust stipe (4.5–6 × 0.8–1.5 cm), and subglobose to ellipsoid basidiospores (5–7.5 × 4–5.5 μm).
Descriptions: Basidioma small to medium-sized. Pileus 6–7.5 cm in diam., plano-convex, margin exceeding lamellae; surface dry, bright yellow to yellow (2A5–7); scales tomentose-fibrillose, dense at the centre, becoming scarce towards the margin, tan (5E6–8) to olive-brown (4E6–8). Lamellae emarginate with decurrent teeth, crowded, edges entire, orange-white to light orange (5A2–4). Stipe 4.5–6 × 0.8–1.5 cm, central, hollow, cylindrical, bright yellow to yellow (2A5–7), with tan (5E6–8) to olive-brown (4E6–8) scales or fibrils.
Basidiospores [60/3/3] 5.5–7.5 × 4–5.5 μm, Q = (1.10–) 1.14–1.63 (−1.75), Qm = 1.35 ± 0.15, subglobose to ellipsoid, in-amyloid, colourless, hyaline, thin-walled, smooth; apiculus relatively small. Basidia 23–30 × 5–7 μm, clavate, 4-spored, hyaline, sterigmata 4.5–5.5 μm long. Pleurocystidia frequent, 32–47 × 5.5–9 μm, cylindrical to narrowly clavate, sometimes with mucronate apex, thin-walled, colourless, hyaline. Cheilocystidia 29–59 × 7–15 μm, narrowly cylindrical, cylindrical to clavate, thin-walled, colourless, hyaline. Pileipellis a cutis with the transition to a trichoderm at regular intervals, composed of 3–9 µm wide, thin-walled, filamentous hyphae, terminal cells 4–11 × 30–57 μm, with round apex. Clamps present at all parts of basidioma.
Known distribution: Solitary to scattered on rotten wood in mixed coniferous broad-leaved forests.
Habitat: North America and East Asia (Holec et al. Citation2019; this study).
Additional material examined: China, Jilin Province, Baishan City, Lushuihe Forest farm, 786 m elev., on rotten wood in a mixed coniferous broadleaved forest, 12 July 2010, HMJAU 23,667; Baishan City, Jinjiang Grand Canyon, 1,049 m elev., on rotten wood in a mixed coniferous broadleaved forest, 19 August , HMJAU 7497.
Notes: Tricholomopsis floccosa was reported as a T. decora complex species (Holec et al. Citation2019) and it is characterised by a bright yellow to yellow pileus with olive-brown scales. It can be distinguished from T. badinensis by its more robust basidioma and shorter pleurocystidia and from T. decora by its darker pileus, coarser scales, more robust basidiomata, and subglobose to ellipsoid basidiospores.
Tricholomopsis sect. Glabrae Zhu L. Yang & G. S. Wang, sect. nov.
Mycobank: MB 849,780.
Type: Tricholomopsis glabra Zhu L. Yang & G. S. Wang.
Diagnosis: Tricholomopsis sect. Glabrae can be distinguished from the other sections of Tricholomopsis by its glabrous pileus, adnate to decurrent lamellae, slender stipe, and the absence of pleurocystidia. Species of sect. Glabrae prefer to live on stumps of bamboo.
Description: Basidioma small to medium-sized, tricholomoid; pileus yellow, glabrous, seldom tomentose; lamellae adnate; stipe clavate, hollow; basidiospores subglobose to ellipsoid, smooth, inamyloid; basidia clavate, 4-spored; cheilocystidia prominent or absent; pleurocystidia absent or present; pileipellis cutis, seldom trichoderm; prefers forests with bamboo.
Notes: To date, four species of sect. Glabrae have been recognised via phylogenetic analyses. Three species in this section have a glabrous pileus, while T. lechatii has a tomentose or velvety pileus.
Key to the species of Tricholomopsis sect. Glabrae in China
(1)Pileus >4 cm in diam; stipe >5 cm in length.................................................................... T. flava
(1) Pileus <4 cm in diam; stipe <5 cm in length ............................................................................ T. glabra
Tricholomopsis flava Zhu L. Yang & G. S. Wang, sp. nov.
Figure 9. Microscopic features of Tricholomopsis flava (type, HKAS 96,940). (a) Basidiospores; (b) Basidia; (c) Pileipellis; (d) Cheilocystidia; (e) Caulocystidia. Bars: a – e = 10 μm. Drawing by Zhu-Liang Yang.
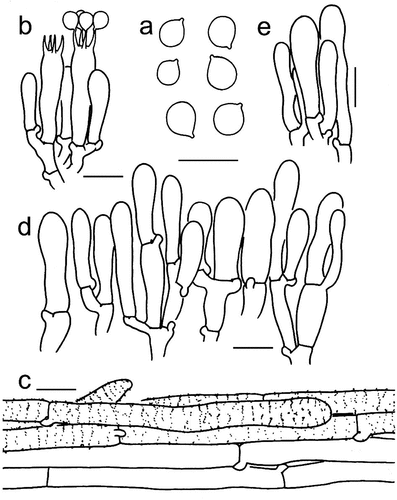
Mycobank: MB 849,781.
Etymology: flava referring to the yellow pileus.
Type: China, Yunnan Province, Puer City, Lancang County, Mengben Village, 1,490 m elev., on stumps of bamboo, 22 August , Yan-Jia Hao 105 (HKAS 96,940, holotype).
Diagnosis: Tricholomopsis flava differs from the other species of sect. Glabrae by its larger pileus (4–9 cm), robust stipe (6–8 × 0.8–1 cm), subdecurrent to decurrent lamellae, and pileipellis cutis.
Description: Basidiomata small to medium-sized. Pileus 4–9 cm in diam., at first convex, plano-concave, sometimes with wavy-irregular margin; surface dry, light brown (4C6–8) at centre with radially light yellow (4A2) to white (2A3) striate, becoming sulphur yellow (2A5–8) towards the margin, surface glabrous. Lamellae subdecurrent to decurrent, crowded, edges even, pale yellow (3A2–3). Stipe 6–8 × 0.8–1 cm, central, cylindrical, hollow, sometimes oblate or curved, sulphur yellow (2A5–8) with longitudinally light yellow (4A2) to white (2A1) striations.
Basidiospores [60/3/3] 4–6 × 4–5 μm, Q = 1–1.25(−1.39), Qm = 1.13 ± 0.09, globose to broadly ellipsoid, inamyloid, colourless, hyaline, thin-walled. Basidia 20–31 × 5–9 μm, cylindrical to clavate, 4-spored, hyaline, sterigmata 2–4 μm long. Pleurocystidia absent. Cheilocystidia frequent, 17–45 × 5–24 μm, cylindrical to clavate. Pileipellis a cutis of parallel cylindrical hyphae 4–9 μm broad, with yellow pigments. Clamps present at all parts of basidioma.
Known distribution: China.
Habitat: Solitary to clustered on stumps of bamboo.
Additional material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Xishuangbanna Botanical Garden, 540 m elev., on stumps of bamboos, 7 August , Qi Zhao Citation2653 (HKAS 90,527); Puer City, Lancang County, 1,490 m elev., on stumps of bamboos, 22 August , Qing Cai 74 (HKAS 129,338).
Notes: Tricholomopsis flava is most phylogenetically similar to T. aurea. However, macro-morphologically, the pileus of T. aurea is smaller and the stipe is shorter than that of T. flava. Micro-morphologically, the cheilocystidia of T. aurea are longer than that of T. flava. Tricholomopsis flava is also similar to T. badinensis, T. decora, and T. sulphureoides, but it can be distinguished from the other three by its glabrous pileus. Furthermore, T. flava has a slenderer stipe, smaller and rounder basidiospores, and shorter cheilocystidia.
Tricholomopsis glabra Zhu L. Yang & G. S. Wang, sp. nov.
Figure 10. Microscopic features of Tricholomopsis glabra (type, HKAS 129,332). (a) Hymenium; (b) Basidiospores; (c) Pileipellis. Bars: a – b = 10 μm; c = 20 μm.
Mycobank: MB 849,782.
Etymology: glabra referring to a glabrous pileus.
Type: China, Anhui Province, Huangshan City, Yungu Temple, 852 m elev., on stumps of bamboo, 15 July 2018, Ting Guo 1067 (HKAS 129,332, holotype).
Diagnosis: Tricholomopsis glabra differs from the other species of T. sect. Glabrae by its yellow-white to pastel yellow glabrous pileus, adnate lamellae, slender stipe (3–4 × 0.2–0.4 cm) presence of pleurocystidia, and absence of cheilocystidia.
Description: Basidiomata small to medium-sized. Pileus 2–4 cm in diam., at first hemispherical, then plano-convex, margin straight to inflexed; surface dry, yellow-white, pastel yellow (3A2–4) to bright yellow (3A6–8), sometimes olive (3E6–8) at the margin, glabrous. Lamellae adnate, crowded, edge entire, yellow-white, pastel yellow (3A2–4) to bright yellow (3A6–8). Stipe 3–4 × 0.2–0.4 cm, cylindrical, hollow, yellow-white, pastel yellow (3A2–4) to bright yellow (3A6–8), glabrous.
Basidiospores [41/2/2] 4–6 × 3.5–5 μm, Q = 1–1.4, Qm = 1.2 ± 0.1, globose, broadly ellipsoid to ellipsoid, inamyloid, colourless, hyaline, thin-walled, smooth; apiculus relatively small. Basidia 17–27 × 4.5–7 μm, cylindrical to clavate, 4-spored, hyaline, sterigmata 3–4 μm long. Pleurocystidia 22–38.5 × 4–6 μm, narrowly cylindrical to flexuose, thin-walled, colourless, hyaline. Cheilocystidia absent. Pileipellis a cutis to trichoderm, composed of 4–12 μm wide, thin-walled, filamentous hyphae, terminal cells cylindrical 5–8 × 20–45 μm. Clamps present at all parts of basidioma.
Known distribution: China.
Habitat: Solitary to scattered on stumps of bamboo.
Additional material examined: China, Hunan Province, Yuelu Mountain, 208 m elev., on stumps of bamboo, 4 January 2017, Ping-Wu Luo (HKAS 129,333).
Notes: Tricholomopsis glabra is characterised by a pale yellow pileus without scales, slender basidiospores, and the absence of cheilocystidia. It has larger basidioma and more slender basidiospores than T. aurea and smaller basidioma and paler pilei than T. flava. Tricholomopsis glabra also lacks cheilocystidia, while T. flava lacks pleurocystidia.
Tricholomopsis sect. Tricholomopsis
Type: Tricholomopsis rutilans (Schaeff.) Singer.
Description: Basidioma tricholomoid; pileus densely covered with purplish to red scales; basidiospores globose to oblong, smooth, inamyloid; cheilocystidia prominent; pileipellis a cutis to a trichoderm; solitary or scattered on rotten wood or soil in coniferous or broad-leaved forests, seldom on grassland.
Notes: Tricholomopsis sect. Tricholomopsis can be distinguished from the other sections by its purplish to red scales. To date, nine taxa of sect. Tricholomopsis have been identified via phylogenetic analyses, including two novel species and seven known taxa.
Key to the species of Tricholomopsis sect. Tricholomopsis in China
(1) Pileus with brownish or darkish scales; stipe <0.5 cm in width.................................................. 2
(1) Pileus with purplish to red scales; stipe >0.5 cm in width............................................................................ 3
(2) Basidiospores broadly ellipsoid to ellipsoid; pleurocystidia <50 μm in length................ T. sulphureoides
(2) Basidiospores ellipsoid to fusiform; pleurocystidia >50 μm........................................................... T. pallidolutea
(3) Scales on pileus mealy, warped; stipe slender, less than 1 cm in width.................................................................. 4
(3) Scales on pileus tomentose, adpressed; stipe robust, more than 1 cm in width...................................................... 5
(4) Pileus depressed; basidiospores wider than 4.5 μm.................................................................. T. depressa
(4) Pileus applanate; basidiospores narrower than 4.5 μm......................................................... T. flammula
(5) On buried litter in meadows; scales dark blond to orange........................................................... T. campestris
(5) On rotten wood; scales red or purple-red............................................................................................... 6
(6)Pleurocystidia absent orscarce....................................................................... T. rutilans
(6)Pleurocystidia present or abundant.................................................................................... 7
(7) Stipes covered with dense scales; scales purple-red............................................................ T. yunnanensis
(7) Stipes covered with scarce scales; scales brown.................................................. T. mitirubicunda
Tricholomopsis campestris Zhu L. Yang & G. S. Wang, sp. nov.
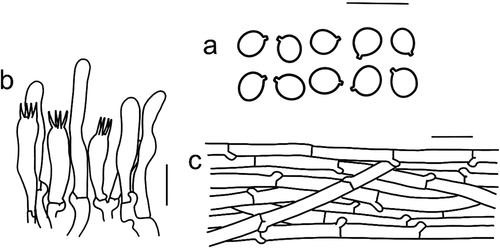
Figure 11. Microscopic features of Tricholomopsis campestris (type, HKAS 116,178). (a) Basidiospores; (b) Hymenium; (c) Pileipellis; (d) Cheilocystidia. Bars: a – b = 10 μm, c – d = 20 μm.
Mycobank: MB 849,777.
Etymology: referring to the habitat of basidioma on buried litter on meadows.
Type: China, Tibet Autonomous Region, Nyingchi, Muduohuahai Scenic Area, 3,475 m elev., on buried litter in meadows, 21 July 2019, Geng-Shen Wang 476 (HKAS 116,178, holotype).
Diagnosis: Tricholomopsis campestris differs from the other species of sect. Tricholomopsis by its castaneous squamose pileus and stipe and stuffed stipe. It is the only species living on buried substrate in meadows above 3,400 m altitude.
Description: Basidiomata small to medium-sized. Pileus 4–8 cm in diam., plano-convex, with margin deflexed; surface dry, pale yellow (2A2–3) to bright yellow (3A3–5), surface densely covered with dark blond (5D4–5), tawny (5D6–8) to orange (6B6–8) tomentose-fibrillose scales or fibrils. Lamellae emarginate with decurrent teeth to subdecurrent, crowded, edges entire, white (1A1), yellow white (1A2–3) to pastel yellow (3A4), yellow (3A5). Stipe 4–5 × 1–1.5 cm, central, solid, cylindrical to slightly tapering upwards, pastel yellow (3A4–7), with dark blond (5D4–5), tawny (5D6–8) to orange (6B6–8) tomentose-fibrillose scales or fibrils.
Basidiospores [44/2/2] 5.5–7.5 × 4–5 μm, Q = 1.20–1.75(−1.80), Qm = 1.42 ± 0.09, broadly ellipsoid to ellipsoid, inamyloid, colourless, hyaline, thin-walled, smooth; apiculus relatively small. Basidia 22–32 × 6–8 μm, cylindrical to clavate, 4-spored, hyaline, sterigmata 6–8 μm long. Pleurocystidia frequent, 30–48 × 6–8 μm, cylindrical, with obtuse or rostrate apex, thin-walled, colourless, hyaline. Cheilocystidia 45–85 × 14–30 μm, narrowly clavate to clavate, thin-walled, colourless, hyaline. Pileipellis a cutis with the transition to a trichoderm at regular intervals, composed of 5–8 µm wide, thin-walled filamentous hyphae, terminal cells 20–45 × 6–13 μm, with round apex. Clamps present at all parts of basidioma.
Known distribution: southwestern China.
Habitat: Solitary on buried litter in meadows.
Additional material examined: China, Yunnan Province, Diqing Tibetan Autonomous Prefecture, Shangri-La, 4,251 m elev., on buried litter in meadow, 8 August , Jian-Wei Liu 495 (HKAS 98,074).
Notes: Tricholomopsis campestris formed a clade sister to T. rutilans. The castaneous scales on the pileus and stipe differentiate it from T. rutilans. In Tricholomopsis, most species have a hollow stipe, while T. campestris has a solid stipe. In addition, T. campestris is also the only known species in Tricholomopsis that lives on buried litter in meadows above 3,400–4,200 m altitude.
Tricholomopsis depressa Zhu L. Yang & G. S. Wang, sp. nov.
Figure 12. Microscopic features of Tricholomopsis depressa (type, HKAS 53,624). (a) Basidiospores; (b) Hymenium; (c) Pileipellis; (d) Cheilocystidia. Bars: a – b = 10 μm, c – d = 20 μm.
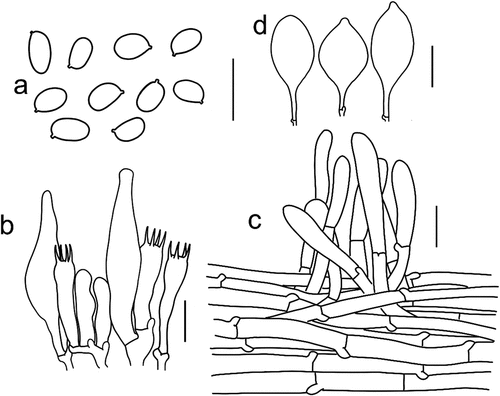
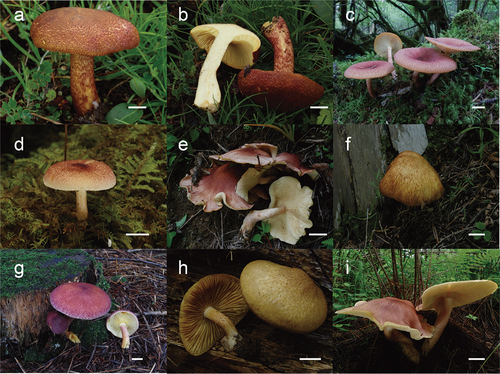
Figure 13. (a – b) Tricholomopsis compestris (HKAS 116,178, photo by Geng-Shen Wang); (c) T. depressa (HKAS 53,624, photo by Zai-Wei Ge); (d) T. flammula (HKAS 116,169, photo by Geng-Shen Wang); (e) T. mitirubicunda (HKAS 71,469, photo by Yan-Jia Hao); (f) T. pallidolutea (HKAS 129,339, photo by Jian-Wei Liu); (g) T. rutilans (HKAS 105,394, photo by Xiao-Xia Ding); (h) Tricholomopsis sulfureoides (HKAS 51,002, photo by Zaiwei Ge); (i) T. yunnanensis (HKAS 76,311, photo by Yanjia Hao). Bars: a – i = 1 cm.
Mycobank: MB 849,778.
Etymology: depressa, from depressus = depressed, referring to a depressed pileus.
Type: China, Tibet Autonomous Region, Garze, Daofu County, 3,000 m elev., on rotten wood in Abies forest, 25 July 2007, Zai-Wei Ge Citation1538 (HKAS 53,624, holotype).
Diagnosis: Tricholomopsis depressa is characterised by its small to medium-sized basidioma, a depressed pileus covered with small, grey pink to pink scales, and a smooth stipe with yellowish to pink scales. It can be distinguished from T. flammula by a robust stipe and depressed pileus, while it can be distinguished from T. rutilans by a smaller and slender basidioma and elongated basidiospores.
Description: Basidioma small to medium-sized. Pileus 2–6 cm in diam., at first hemispherical to convex, then plano-convex, often depressed at centre, margin involute; surface dry, cream (3A2), yellowish (3A3–4) to grey yellow (3B5–7), densely covered with small, grey pink (11B4–6) to pink (12A3–6) scales. Lamellae adnate with decurrent teeth, crowded, edges entire, yellow-white (2A2–4) to yellowish (3A2–3). Stipe 2–7 × 0.5–1.2 cm, cylindrical, sometimes slightly tapering downwards, hollow, smooth or with yellow white (2A2–4) to yellowish (3A2–3), with grey pink (11B4–6) to pink (12A3–6) scales or fibrils.
Basidiospores [64/3/3] 5.5–7.5 × 3.5–5.0 μm, Q = (1.33–) 1.38–2.00, Qm = 1.64 ± 0.16, ellipsoid to elongate, in-amyloid, colourless, hyaline, thin-walled, smooth; apiculus relatively small. Basidia 21–36 × 5.5–8.5 μm, clavate, 4-spored, hyaline, sterigmata 2–4 μm long. Pleurocystidia frequent, 25–70 × 7–26 μm, ventricose-subfusiform, thin-walled, colourless, hyaline. Cheilocystidia 50–78 × 16–29 μm, clavate to spheropedunculate, apex sometimes mucronate, thin-walled, colourless, hyaline. Pileipellis a cutis with the transition to a trichoderm at regular intervals, composed of 5–11 µm wide, thin-walled, filamentous hyphae, terminal cells subcylindrical to cylindrical, 40–69 × 8–12 μm, with round apex. Clamps present at all parts of basidioma.
Known distribution: China and the Czech Republic.
Habitat: Solitary to scattered on rotten wood in Abies forests.
Additional material examined: China, Tibet Autonomous Region, Nyingchi, Milin Village, 2,950 m elev., on rotten wood in Pinus forest, 29 July 2014, Qi Zhao Citation2118 (HKAS 87,884); Riwoqe, Mengda Village, 4,100 m elev., on rotten wood in Picea forest, 9 August , Zai-Wei Ge 312 (HKAS 46,092); same locality, altitude 3900 m, on rotten wood in Picea forest, 10 August , Zai-Wei Ge 317 (HKAS 46,097); Riwoqe, Sangduo Village, 3900 m elev., on rotten wood in Picea forest, 11 August , Zhu-Liang Yang Citation4364 (HKAS 45,743). Sichuan Province, Xiaojin County, Shalong Village, 2723 m elev., on rotten wood in Abies forest, 26 July 2007, Zai-Wei Ge1545 (HKAS 53,631). Gansu Province, Gannan City, Diebu, 3,000 m elev., on rotten wood in Abies forest, 12 August , Xue-Tai Zhu 693 (HKAS 76,542). Yunnan Province, Zhongdian County, 3,500 m elev., on rotten wood in Picea forest, 25 July 2006, Yan-Chun Li 633 (HKAS 50,387); same locality, 3,600 m elev., on rotten wood in Picea forest, 7 August , Zhu-Liang Yang Citation4560 (HKAS 48,724). Jilin Province, Xifeng County, 207 m elev., on rotten wood, 2 August , Yan-Chun Li Citation1186 (HKAS 56,040); Chibei District, Dayangcha Farmed Forest, 860 m elev., on rotten wood, 2 August , Zhu-Liang Yang Citation5101 (HKAS 54,397); Antu County, Erdaobaihe, Changbai Mountains Nature Reserve, Baishan Station, 490 m elev., on rotten wood in mixed forests with Pinus, Abies, Populus, and Betula, 10 September 2006, Xiang-Hua Wang Citation2091 (HKAS 51,808).
Notes: One collection of Tricholomopsis depressa was once identified as T. flammula (Holec and Kolařík Citation2013). With additional analyses, we confirmed that T. depressa was a monophyletic clade sister to T. flammula and can be differentiated by its larger basidiomata and depressed pileus.
Tricholomopsis flammula Métrod ex Holec, J. National Mus. (Prague), Nat. Hist. Ser. 178: 8 (2009)
Known distribution: Europe, East Asia, and North America (Métrod Citation1946; Smith Citation1960; Krisai-Greilhuber and Voglmayr Citation2000; Holec Citation2009; Holec and Kolařík Citation2011, Citation2013; Razaq et al. Citation2012; Wang et al. Citation2018; this study).
Habitat: Solitary to scattered on decaying wood chips or sawdust remnants in coniferous or broad-leaved forests.
Material examined: China, Hubei Province, Shennongjia, 2600 m elev., on rotten wood, 1 August , Xiang-Hua Wang Citation2494 (HKAS 73,509). Sichuan Province, Baiyu County, Marong Town, 3490 m elev., on rotten wood, 20 August , Zai-Wei Ge Citation1340 (HKAS 50,927). Tibet Autonomous Region, Nyingchi, Lulang Town, 3,400 m elev., on rotten wood in coniferous broad-leaved forests, 20 July 2019, Geng-Shen Wang 467 (HKAS 116,169). Yunnan Province, Dali Bai Autonomous Prefecture, Binchuan County, Jizu Mountain, 743 m elev., on rotten wood, 4 August , Jiao Qin 743 (HKAS 81,181).
Notes: Tricholomopsis flammula was first described in France by Métrod (Citation1946) and then validly published by Holec (Citation2009). It differs from T. rutilans by its smaller and much slender basidiocarps, smaller and finer scales, elongated basidiospores, and very abundant pleurocystidia.
Tricholomopsis mitirubicunda L. Fan & N. Mao, in Mao, Xu & Fan, Phytotaxa 507(2): 161 (2021)
Known distribution: Europe and East Asia (Olariaga et al. Citation2015; Mao et al. Citation2021; this study).
Habitat: Solitary to scattered on rotten wood or buried wood in coniferous or broad-leaved forests.
Material examined: China, Hubei Province, Shennongjia Forestry District, Lishubao Town, 1,324 m elev., on buried debris in Pinus forest, 14 July 2012, Jiao Qin 513 (HKAS 77,914); Shennongjia Forestry District, Hongping Town, 1,900 m elev., on buried debris in Pinus forest, 15 July 2012, Jiao Qin 538 (HKAS 77,939); Shiyan City, Maojian District, 320 m elev., on buried debris in a forest with Pinus and Quercus, 1 July 2013, Qi Zhao Citation1842 (HKAS 80,952). Shandong Province, Taian City, Taishan Mountain, 982 m elev., on buried debris in Pinus forest, 4 August , Yan-Chun Li Citation2805 (HKAS 89,448). Yunnan Province, Baoshan City, Longyang County, 1,684 m elev., on buried debris in Pinus forest, 10 January 2003, Yan-Chun Li Citation1739 (HKAS 59,486); Zhaotong City, 1,900 m elev., on buried debris in Pinus forest, 17 August , Si-Peng Jian 71 (HKAS 101,060); Kunming City, Yeyahu, 2,100 m elev., in Fagaceae forest, 3 September 2017, Sai Gong 20 (HKAS 101,376); Lushui County, Pianma, 1,400 m elev., on rotten wood in a forest with Fagaceae and Theaceae, 5 August , Yan-Jia Hao 360 (HKAS 71,469); Dali Bai Autonomous Prefecture, Cang Mountains, 2,900 m elev., on buried debris in coniferous forests, 22 August , Gang Wu 116 (HKAS 57,648). Anhui Province, Huoshan County, Taiyang Village, 600 m elev., on buried debris in a forest with Pinus and Fagaceae, 27 June 2013, Yan-Jia Hao 882 (HKAS 80,162); Huoshan County, Taiyang village, 446 m elev., on buried debris in Pinus forest, 27 June 2013, Ting Guo 651 (HKAS 81,853). Jilin Province, Changchun City, Jingyuetan Park, 350 m elev., on buried debris in a broadleaved and coniferous forest, 5 September 2015, Jing Li 376 (HKAS 91,478). AUSTRIA, Kleinwalsertal, 1,100 m elev., on rotten wood, 26 September 2016, Zhu-Liang Yang Citation5915 (HKAS 129,326).
Notes: Tricholomopsis mitirubicunda was regarded as “T. aff. rutilans 2” by Olariaga et al. (Citation2015) and later published by Mao et al. (Citation2021). It differs from T. rutilans by the presence of abundant pleurocystidia (Mao et al. Citation2021).
Tricholomopsis pallidolutea L. Fan & N. Mao, in Mao, Xu & Fan, Phytotaxa 507(2): 160 (2021)
Known distribution: northwestern China (Mao et al. Citation2021; this study).
Habitat: solitary or scattered on trunks or buried wood in forests with Picea or Larix.
Material examined: China, Xinjiang Uygur Autonomous Region, Ili Kazakh Autonomous Prefecture, Zhaosu County, 1,886 m elev., solitary on soil in a forest with Picea, 14 July 2020, Jian-Wei Liu 752 (HKAS 129,339).
Notes: Tricholomopsis pallidolutea, T. sulphureoides, and T. flavescens are three species with brown fibrillose pileus scales. However, T. pallidolutea can be distinguished from T. sulphureoides and T. flavescens by its more elongated basidiospores and shorter pleurocystidia.
Tricholomopsis rutilans (Schaeff.) Singer, Schweiz. Z. Pilzk. 17: 56 (1939)
Known distribution: Europe, North America, East Asia, and Australia (Smith Citation1960; Liu Citation1994; Holec and Kolařík Citation2013; Olariaga et al. Citation2015; Cooper and Park Citation2016).
Habitat: Solitary to scattered on rotten wood or buried wood in coniferous or broad-leaved forests.
Material examined: China, Sichuan Province, Luoji Town, Luoji Mountain, 2,000 m elev., on rotten wood in forests with Pinus, 28 July 2012, Yan-Jia Hao 657 (HKAS 76,315). Yunnan Province, Ninglang County, Zhanhe Country, 3,000 m elev., on rotten wood in mixed forests with Quercus, Pinus, and Rhododendron, 17 July 2010, Qi Zhao 786 (HKAS 69,507); Yulong County, Gaomeigu, 3,300 m elev., on rotten wood, 20 July 2008, Qi Zhao 835 (HKAS 55,036); Yulong County, Yulong Reservoir, 3,100 m elev., on rotten wood, 29 July 2006, Yan-Chun Li 674 (HKAS 50,428); Lijiang City, Yulong Mountain, 2,800 m elev., in a forest with Pinus, 1 August , Mu Zang 10,208 (HKAS14979). Tibet Autonomous Region, Zuogong County, Wangda Town, 3,900 m elev., on rotten wood, 18 July 2009, Zhu-Liang Yang Citation5287 (HKAS 57,830). Liaoning Province, Dandong City, Kuandian County, 500 m elev., on buried debris in a coniferous forest, 29 August , Xiao-Xia Ding 384 (HKAS 105,394). Heilongjiang Province, Yichun City, Dailing Town, 225 m elev., on rotten wood, 15 August , Xiang-Hua Wang Citation2631 (HKAS 61,828).
Notes: Tricholomopsis rutilans s. str. was designated a lectotype and an epitype by Olariaga et al. (Citation2015). It is similar to T. mitirubicunda and T. yunnanensis. But T. rutilans has no or only scarce pleurocystidia, while T. mitirubicunda and T. yunnanensis have abundant pleurocystidia. The basidiospores of T. mitirubicunda are much more slender than those of T. yunnanensis. The T. rutilans is nearly distributed cosmopolitan, while T. mitirubicunda is distributed in Europe and East Asia, while T. yunnanensis is only known from southwestern China.
Tricholomopsis sulphureoides (Peck) Singer, Annls mycol. 41(1/3): 69 (1943)
Known distribution: Europe, Asia, and North America (Smith Citation1960, Vauras 2009; Holec and Kolařík Citation2011, Citation2013; Holec Citation2012; Saar and Voitk Citation2015; this study).
Habitat: Solitary to scattered on rotten wood in coniferous or broad-leaved forests.
Material examined: China, Sichuan Province, Litang County, Junba Village, 4,280 m elev., on rotten wood, 26 August , Zai-Wei Ge Citation1416 (HKAS 51,002).
Notes: Tricholomopsis sulphureoides was first described in the United States as Agaricus sulfuroides by (Peck Citation1872). Phylogenetic analysis revealed that T. sulphureoides and T. flavescens formed two independent clades, although distinct morphological characteristics between them were not observed in the study by Saar and Voitk (Citation2015). Tricholomopsis sulphureoides can be easily distinguished from T. flammula, T. rutilans, T. depressa, T. yunnanensis, and T. pteridiicola by minute brown or absent scales.
Tricholomopsis yunnanensis (M. Zang) Li R. Liu, Yan C. Li & Zhu L. Yang, in Liu, Wang, Jia, Kang, Yang & Li, Phytotaxa 530(2): 184 (2022)
Synonym: Tricholomopsis galeata L. Fan & N. Mao, in Mao, Xu & Fan, Phytotaxa 507 (2): 160 (2021).
Known distribution: China (Zang and Zeng Citation1978; Mao et al. Citation2021; Liu et al. Citation2022).
Habitat: Solitary to scattered on rotten wood or buried wood in the forests of Pinus.
Material examined: China, Yunnan Province, Kunming City, Heilongtan Park, 2,700 m elev., on rotten wood, 2 September 1978, You-Feng Yu 3884 (HKAS 40,151); same locality, on rotten wood, 8 August , Wen-Qing Yin 931 (HKAS 28,976); same locality, on buried debris in Pinus forest, 9 August , Geng-Shen Wang 194 (HKAS 109,582); Kunming Botanical Garden, 1,990 m elev., on buried debris in Pinus forest, 18 August , Geng-Shen Wang Citation1335 (HKAS 121,974); Yeyahu, 2,100 m elev., on buried debris in Pinus forest, 1 September 2013, Zhu-Liang Yang Citation5757 (HKAS 80,034); Nujiang City, Lanping County, Xihe Village, 2,760 m elev., on buried debris in Pinus forest, 15 August , Qi Zhao 890 (HKAS 69,611); Chuxiong Yi Autonomous Prefecture, Nanhua County, Zixi Mountain, 2,250 m elev., on buried debris in Pinus forest, 12 July 2001, Xiang-Hua Wang Citation1230 (HKAS 39,166); Dali Bai Autonomous Prefecture, Yongping County, Longmen Village, 2,282 m elev., on buried debris in Pinus forest, 31 July 2009, Qing Cai 65 (HKAS 58,732); Nanjian County, 2,317 m elev., on buried debris in Pinus forest, 27 July 2009, Li-Ping Tang 965 (HKAS 56,922); Binchuan County, Jizu Mountain, on buried debris in Pinus forest, 2,450 m elev., 4 August , Jiao Qin 731 (HKAS 81,169); Lijiang City, Xiangshan Mountain, 2,600 m elev., on buried debris in Pinus forest, 5 August , Yan-Chun Li 322 (HKAS 48,555). Sichuan Province, Xichang City, Jiluo Mountain, 2,000 m elev., on buried debris in a forest with broad-leaved and coniferous forest, 18 July 1992, Pei-Qiong Sun Citation1818 (HKAS25752); Puge County, Luoqi Mountain, 2,000 m elev., on buried debris in Pinus forest, 28 July 2012, Yan-Jia Hao 653 (HKAS 76,311); same locality, on buried debris in Pinus forest, 28 July 2012, Kuan Zhao 18 (HKAS 77,370). Shandong Province, Qingzhou City, Yangtianshan National Forest Park, 520 m elev., on buried debris in Pinus forest, 22 August , Xiang-Hua Wang Citation3008 (HKAS 73,557).
Notes: Tricholomopsis yunnanensis was first described in southwestern China as Paxillus yunnanensis by Zang and Zeng (Citation1978). The type was restudied by Liu et al. (Citation2022), who placed it into Tricholomopsis as T. yunnanensis. Phylogenetic analysis indicated that T. yunnanensis and T. galeata are conspecific. Although Mao et al. (Citation2021) did not find any pleurocystidia in T. galeata and Liu et al. (Citation2022) regarded the pileipellis as palisadoderm, we observed the presence of pleurocystidia and regarded the pileipellis as a cutis with transition to a trichoderm in T. yunnanensis.
Tricholomopsis sect. Lividae A. H. Smith, Brittonia 12(1): 57 (1960)
Type: Tricholomopsis totilivida (Murrill) Singer.
Notes: This section contains only one species and can be easily distinguished from the other sections by its livid pileus and lamellae and its large pleurocystidia. It has only been known from Florida, USA. It may be close to T. sect. Tricholomopsis due to its squamous pileus. However, this section needs further research and molecular data to verify its position within Tricholomopsis.
Other species
Tricholomopsis lividipileata P. G. Liu, Acta Mycol. Sin. 13(3): 182 (1994)
Known distribution: China (Liu Citation1994).
Habitat: scattered or clustered in litterfall in forests with Pinus.
Notes: Tricholomopsis lividipileata was described in Sichuan, China, by Liu (Citation1994), and it is an edible species favoured by local people (Yuan & Sun Citation2007). However, subsequent visits by our expedition team to Sichuan failed to identify this species. The descriptions and illustrations of T. lividipileata are very similar to those of Tricholoma olivaceum by Rambaut et al. (Citation2018). Both of them have an olive-brown pileus with fibrils, similar basidiospores, and cutis pileipellis, and they live in coniferous forests at similar altitudes. These clues suggested that T. lividipileata may belong to Tricholoma and be close to Ta. olivaceum. Whether they are conspecific remains an open question, because the type of T. lividipileata is missing.
Tricholomopsis shulanensis X. He, Acta Mycol. Sinica 8(3): 202 (1989)
Known distribution: China (He Citation1989).
Habitat: Single or scattered, on rotting sticks buried in soil under hazel wood.
Notes: Specimens of Tricholomopsis shulanensis were not traced and studied in this study. Judging from the protologue, T. shulanensis is very similar to T. flammula with subtle differences. The identity of this species needs to be verified using molecular data in the future.
Tricholoma nigrosquamosum (P. G. Liu) Zhu L. Yang & G. S. Wang, comb. nov.
Mycobank: MB 849,786.
Basionym: Tricholomopsis nigrosquamosa P. G. Liu, Acta Mycol. Sinica 13(3): 183 (1994).
Synonymy: Tricholoma sinopardinum Zhu L. Yang, X. X. Ding, G. Kost & Rexer, Phytotaxa 305(1): 6 (2017).
Known distribution: China (Liu Citation1994).
Habitat: Scattered under litter in forests with Abies.
Material examined: China, Sichuan Province, Rang Tang, 3,500 m elev., scattered under litterfall in a forest with Abies, 28 July 1991, Ming-Sheng Yuan 1434 (HKAS 23,947, type of T. nigrosquamosum). Tibet Autonomous Region, Jiangda County, Jiangda Town, 3,500 m elev., in forest dominated by Picea sp. and Populus sp., 8 August , Bang Feng 1427 (HKAS 82,533, type of Ta. sinopardinum).
Notes: Tricholomopsis nigrosquamosum was described from Sichuan by Liu (Citation1994), while Ta. sinopardinum Zhu L. Yang et al. was described from Tibet by Yang et al. (Citation2017). The type of T. nigrosquamosum (HKAS 23,947) was sequenced, and the ITS identity between T. nigrosquamosum and Ta. sinopardinum was 100%. Morphologically, they are the same. Therefore, the new combination, Ta. nigrosquamosum is proposed.
Sarcomyxineae Zhu L. Yang & G. S. Wang, subord. nov.
MycoBank: MB 849,775.
Type genus: Sarcomyxa P. Karst. 1891.
Etymology: From the type genus Sarcomyxa.
Diagnosis: Sarcomyxineae differs from the other suborders of Agaricales by its lignicolous habitat; fan- or kidney-shaped, viscid pileus with a pellicle easily peeling off from the context; lateral short stipe; prominent cheilocystidia; small allantoid to subcylindrical amyloid basidiospores; and the common presence of clamp connections.
Description: Basidioma pleurotoid, small to large, fan-or kidney-shaped, fleshy. Pileus viscid or velutinous; pellicle easily peeling off from the context. Lamellae decurrent, crowded. Stipe lateral, short, subcylindrical, floccose. Basidiospores allantoid or cylindrical, thin-walled, colourless and hyaline, smooth, amyloid; apiculus indistinct or very small. Basidia clavate, mostly 4-spored, thin-walled. Cheilocystidia clavate. Pleurocystidia clavate to nearly fusiform. Pileipellis a gelatinised plagiotrichoderm. Clamp connections present in all parts of basidioma.
Known distribution: Europe, North America, and Asia.
Habitat: Saprotrophic, on rotten wood in coniferous or broad-leaved forests.
Family included: Sarcomyxaceae Olariaga et al.
Notes: Sarcomyxaceae was once placed in the suborder Pleurotineae by Olariaga et al. (Citation2020). Our phylogenomic analyses showed that Sarcomyxaceae formed a unique suborder clade ( and 2). Due to its unique phylogenetic position and morphological features, it is considered an independent suborder here. To date, four species have been described for the genus Sarcomyxa (Cai et al. Citation2023).
Acknowledgments
The authors are very grateful to Drs. B. Feng, H. Qu, S.Z. Zhou, Z.M. He, and T. James for providing kind help and polishing the English of the first version of this work. The genome sequencing of Typhula sp., Bolbitius vitellinus, Conocybe apala, Cystostereum murrayi, Favolaschia claudopus, Flagelloscypha sp., Leratiomyces erythrocephalus, Lycoperdon perlatum, Mycocalia denudate, Panellus stipticus, Phaeosolenia platensis, Radulomyces confluens, and Schizophyllum radiatum was conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. We thank Drs. F. Martin, R. Cazenave, L.G. Nagy, A. Nilsen, D. Catcheside, H.L. Liao, A. Rojas, T. Rämä, S. Maurice, O. Miettinen, I. Skrede, H. Kauserud, A. Kohler, I. Choi, J. Magnuson, R.A. Ohm and G. Bonito for allowing us to use their unpublished genomes in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5): 455–477. doi: 10.1089/cmb.2012.0021.
- Bánki O, Roskov Y, Döring M, Ower G, Vandepitte L, Hobern D, Remsen D, Schalk P, DeWalt RE, Keping M, et al. 2022. Catalogue of life checklist (annual checklist 2022). Catalogue of Life. doi:10.48580/dfq8
- Cai Q, Lin W-F, Liu J-W, Zhou Y, Yang ZL. 2023. Two new species of Sarcomyxa (Agaricales) from China. Mycol Prog. 22(6):38. doi: 10.1007/s11557-023-01883-8.
- Cai Q, Tulloss RE, Tang LP, Tolgor B, Zhang P, Chen ZH, Yang ZL. 2014. Multi-locus phylogeny of lethal amanitas: implications for species diversity and historical biogeography. BMC Evolution Biology. 14(1):143. doi: 10.1186/1471-2148-14-143.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi: 10.1093/bioinformatics/bty560.
- Cooper JA, Park D. 2016. The fungal genus Tricholomopsis (Agaricales) in New Zealand, including Tricholomopsis scabra sp. nov. Phytotaxa. 288(1):69–70. doi: 10.11646/phytotaxa.288.1.7.
- Dentinger BTM, Gaya E, O’Brien H, Suz LM, Lachlan R, Díaz-Valderrama JR, Koch RA, Aime MC. 2016. Tales from the crypt: genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol J Linn Soc. 117(1):11–32. doi: 10.1111/bij.12553.
- Desjardin D. 2017. The gymnopoid fungi (Basidiomycota, Agaricales) from the Republic of São Tomé and Príncipe, West Africa. Mycosphere. 8(9):1317–1391. doi: 10.5943/mycosphere/8/9/5.
- Edler D, Klein J, Antonelli A, Silvestro D, Matschiner M. 2021. RaxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML.Matschiner M, editor. Methods Ecology Evol. 12(2):373–377. doi: 10.1111/2041-210X.13512.
- Garrido N. 1988. Agaricales s.l. und ihre Mykorrhizen in den Nothofagus-Wäldern Mittelchiles. Bibliotheca Mycologica. 120:1–528.
- Han LH, Wu G, Horak E, Halling RE, Xu J, Ndolo EST, Sato H, Fechner N, Sharma YP, Yang ZL. 2020. Phylogeny and species delimitation of strobilomyces (Boletaceae), with an emphasis on the Asian species. persoonia. 44(1):113–139. doi: 10.3767/persoonia.2020.44.05.
- He X. 1989. A new species of the genus Tricholomopsis. Acta Mycol Sin. 8(3):202–204. doi: 10.13346/j.mycosystema.1989.03.007.
- Holec J. 2009. Valid publication of the name Tricholomopsis flammula (fungi, Basidiomycota, Tricholomataceae), a species clearly separated from T. rutilans. J Nat Mus Prague Nat Hist Ser. 178(3):7–13.
- Holec J, Kolařík M. 2011. Tricholomopsis flammula (Basidiomycota, Agaricales)—molecular taxonomy, delimitation, variability and ecology. Mycol Prog. 10(1):93–99. doi: 10.1007/s11557-010-0679-0.
- Holec J, Kolařík M. 2013. Tricholomopsis in Europe — phylogeny, key, and notes on variability. Mycotaxon. 121(1):81–92. doi: 10.5248/121.81.
- Holec J, Kunca V, Kolařík M. 2019. Tricholomopsis badinensis sp. nov. And T. sulphureoides—two rare fungi of European old-growth forests. Mycol Prog. 18(3):321–334. doi: 10.1007/s11557-018-1449-7.
- Hongo T. 1959. Notes on Japanese larger fungi (14). J Jpn Bot. 34:239–346.
- Hongo T. 1960. Notes on Japanese larger fungi (15). J Jpn Bot. 35:83–90.
- Hongo T. 1966. Notulae mycologicae (5). Vol. 16. Memoirs of the Faculty of Liberal Arts and Education, Shiga University; pp. 57–62.
- Horak E. 1971. A contribution towards the revision of the Agaricales (fungi) from New Zealand. NZ J Bot. 9(3):402–462. doi: 10.1080/0028825X.1971.10430193.
- Horak E. 1980. Fungi, Basidiomycetes. Agaricales y gasteromycetes secotioides. Flora Criptogámica de Tierra del Fuego. 11(6):1–524.
- Hosen MI, Xu JY, Li T, Gates G, Li TH. 2020. Tricholomopsis rubroaurantiaca, a new species of Tricholomataceae from southern China. Mycoscience. 61(6):342–347. doi: 10.1016/j.myc.2020.06.005.
- Hughes KW, Petersen RH, Mata JL, Psurtseva NV, Kovalenko AE, Morozova OV, Lickey EB, Blanco JC, Lewis DP, Nagasawa E, et al. 2007. Megacollybia (Agaricales). Report Tottori Mycological Institute. 45:1–57.
- Jayawardena RS, Hyde KD, Wang S, Sun Y-R, Suwannarach N, Sysouphanthong P, Abdel-Wahab MA, Abdel-Aziz FA, Abeywickrama PD, Abreu VP, et al. 2022. Fungal diversity notes 1512–1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 117(1): 1–272. doi: 10.1007/s13225-022-00513-0.
- Junier T, Zdobnov EM. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the unix shell. Bioinformatics. 26(13):1669–1670. doi: 10.1093/bioinformatics/btq243.
- Kalichman J, Kirk PM, Matheny PB. 2020. A compendium of generic names of agarics and Agaricales. Taxon. 69(3):425–447. doi: 10.1002/tax.12240.
- Kasuya T, Omori M, Kobayashi K, Hanawa S. 2015. Three species of Agaricales, new records from Ibaraki prefecture. Bulletin Ibaraki Nat Mus. 18:53–56.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 29(2):170–179. doi: 10.1007/BF02100115.
- Kishino H, Miyata T, Hasegawa M. 1990. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol. 31(2):151–160. doi: 10.1007/BF02109483.
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. 3rd ed. London: Eyre Methuen.
- Krisai-Greilhuber I, Voglmayr H. 2000. Tricholomopsis flammula of upper Austria. Beitr Naturk Oberösterreichs. 9:701–704.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Leal-Dutra CA, Griffith GW, Neves MA, McLaughlin DJ, McLaughlin EG, Clasen LA, Dentinger BT. 2020. Reclassification of Pterulaceae Corner (Basidiomycota: Agaricales) introducing the ant-associated genus Myrmecopterula gen. nov., Phaeopterula Henn. and the corticioid Radulomycetaceae fam. nov. IMA Fungus. 11(1). doi: 10.1186/s43008-019-0022-6.
- Li J, Han L, Liu X, Zhao Z, Yang ZL. 2020. The saprotrophic Pleurotus ostreatus species complex: late Eocene origin in East Asia, multiple dispersal, and complex speciation. IMA Fungus. 11(1):10. doi: 10.1186/s43008-020-00031-1.
- Liu PG. 1994. Classification of the genus Tricholomopsis from the southwestern China. Acta Mycol Sin. 13(3):181–187. doi: 10.13346/j.mycosystema.1994.03.005.
- Liu LR, Wang GS, Jia LK, Kang JQ, Yang ZL, Li YC. 2022. Type studies on two paxillus species (paxillaceae, Boletales) described from China. Phytotaxa. 530(2):177–188. doi: 10.11646/phytotaxa.530.2.4.
- Lodge DJ, Padamsee M, Matheny PB, Aime MC, Cantrell SA, Boertmann D, Kovalenko A, Vizzini A, Dentinger BTM, Kirk PM, et al. 2014. Molecular phylogeny, morphology, pigment chemistry and ecology in Hygrophoraceae (Agaricales). Fungal Divers. 64(1): 1–99. doi: 10.1007/s13225-013-0259-0.
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM, Kelley J. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of Eukaryotic, prokaryotic, and viral genomes.Kelley J, editor. Mol Biol Evol. 38(10):4647–4654. doi: 10.1093/molbev/msab199.
- Mao N, Xu YY, Fan L. 2021. Three new species of Tricholomopsis (incertae sedis) from North China based on morphological and molecular data. Phytotaxa. 507(2):155–166. doi: 10.11646/phytotaxa.507.2.3.
- Matheny PB, Curtis JM, Hofstetter V, Aime MC, Moncalvo J-M, Ge Z-W, Yang Z-L, Slot JC, Ammirati JF, Baroni TJ, et al. 2006. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia. 98(6): 982–995. doi: 10.1080/15572536.2006.11832627.
- Métrod G. 1946. Champignons du Jura. Revue de Mycologie (Paris). 11(2–3):74–81.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R, Teeling E. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era.Teeling E, editor. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Mirarab S, Reaz R, MdS B, Zimmermann T, Swenson MS, Warnow T. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 30(17):i541–i548. doi: 10.1093/bioinformatics/btu462.
- Moncalvo J-M, Vilgalys R, Redhead SA, Johnson JE, James TY, Catherine Aime M, Hofstetter V, Verduin SJW, Larsson E, Baroni TJ, et al. 2002. One hundred and seventeen clades of euagarics. Mol Phylogenet Evol. 23(3): 357–400. doi: 10.1016/S1055-7903(02)00027-1.
- Mou GF, Bau T. 2021. Asproinocybaceae fam. nov. (Agaricales, agaricomycetes) for accommodating the genera asproinocybe and Tricholosporum, and description of asproinocybe sinensis and tricholosporum guangxiense sp. nov. J Fungus. 7(12):1086. doi: 10.3390/jof7121086.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Sweden: Uppsala UniversityEvolutionary Biology Centre.
- Olariaga I, Huhtinen S, Læssøe T, Petersen JH, Hansen K. 2020. Phylogenetic origins and family classification of typhuloid fungi, with emphasis on Ceratellopsis, Macrotyphula and Typhula (Basidiomycota). Stud Mycology. 96:155–184. doi:10.1016/j.simyco.2020.05.003.
- Olariaga I, Huhtinen S, Læssøe T, Petersen JH, Hansen K, Parra LA. 2022. (2874–2877) proposals to conserve the names Typhula with a conserved type, Macrotyphula against Sclerotium, and Phyllotopsidaceae against Sclerotiaceae, and to reject the name sclerotium fulvum (Basidiomycota: agaricales). Taxon. 71(2):468–470. doi: 10.1002/tax.12697.
- Olariaga I, Laskibar X, Holec J. 2015. Molecular data reveal cryptic speciation within Tricholomopsis rutilans: description of T. pteridicola sp. nov. associated with pteridium aquilinum. Mycol Prog. 14(4):21. doi: 10.1007/s11557-015-1040-4.
- Paradis E, Schliep K, Schwartz R. 2019. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R.Schwartz R, editor. Bioinformatics. 35(3):526–528. doi: 10.1093/bioinformatics/bty633.
- Peck CH. 1872. Report of the Botanist [1869]. Ann Report NY Mus Nat Hist. 23:27–135.
- Pegler DN. 1977. A preliminary agaric flora of East Africa. Kew Bull. 6:1–615.
- Pegler DN. 1983. Agaric flora of the lesser antilles. Kew Bull. 9:1–668.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA, Susko E. 2018. Posterior summarization in bayesian phylogenetics using Tracer 1.7.Susko E, editor. Syst Biol. 67(5):901–904. doi: 10.1093/sysbio/syy032.
- Razaq A, Khalid AN, Ilyas S. 2012. Tricholomopsis flammula métrod ex Holec (Basidiomycota, Agaricales)-an addition to the mushroom flora of Pakistan based on molecular identification. Pak J Bot. 44:413–416.
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things): phytools: R package. Methods Ecology Evol. 3(2):217–223. doi: 10.1111/j.2041-210X.2011.00169.x.
- Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Saar I, Voitk A. 2015. Type studies of two Tricholomopsis species described by Peck. Mycol Prog. 14(7):46. doi: 10.1007/s11557-015-1068-5.
- Sato H, Tanabe AS, Toju H. 2017. Host shifts enhance diversification of ectomycorrhizal fungi: diversification rate analysis of the ectomycorrhizal fungal genera strobilomyces and afroboletus with an 80‐gene phylogeny. New Phytol. 214(1):443–454. doi: 10.1111/nph.14368.
- Shen XX, JL S, Rokas A. 2021. Dissecting incongruence between concatenation- and quartet-based approaches in phylogenomic data.Brown J, editor. Syst Biol. 70(5):997–1014. doi: 10.1093/sysbio/syab011.
- Shen S, Liu S-L, Zhou L-W. 2023. Taxonomy of Hyphodermella: a case study to show that simple phylogenies cannot always accurately place species in appropriate genera. IMA Fungus. 14(1):11. doi: 10.1186/s43008-023-00116-7.
- Shimodaira H, Goldman N. 2002. An approximately unbiased test of phylogenetic tree selection.Goldman N, editor. Syst Biol. 51(3):492–508. doi: 10.1080/10635150290069913.
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of Log-Likelihoods with applications to phylogenetic inference. Mol Biol Evol. 16(8):1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201.
- Singer R. 1939. Phylogenie und taxonomie der Agaricales. Schweizerische: Zeitschrift für Pilzkunde.
- Singer R. 1943. Type studies on basidiomycetes. II. Mycologia. 35(2):142–163. doi: 10.1080/00275514.1943.12017472.
- Singer R. 1951. The Agaricales in modern taxonomy. Lilloa. 22:1–832.
- Singer R. 1953. Type studies on basidiomycetes. VI. Lilloa. 26:57–159.
- Singer R. 1989. New taxa and new combinations of Agaricales (diagnoses fungorum novorum agaricalium 4). Fieldiana Bot. 21:1–133.
- Singer R, Ovrebo CL, Halling RE. 1990. New species of phylloporus and Tricholomopsis from Colombia, with notes on phylloporus boletinoides. Mycologia. 82(4):452. doi: 10.1080/00275514.1990.12025908.
- Smith AH. 1960. Tricholomopsis (Agaricales) in the Western Hemisphere. Brittonia. 12(1):41–70. doi: 10.2307/2805334.
- Steenwyk JL, Buida TJ, Labella AL, Li Y, Shen X-X, Rokas A, Schwartz R. 2021. PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data.Schwartz R, editor. Bioinformatics. 37(16):2325–2331. doi: 10.1093/bioinformatics/btab096.
- Strimmer K, Rambaut A. 2002. Inferring confidence sets of possibly misspecified gene trees. Proc R Soc B Biol Sci. 269(1487):137–142. doi: 10.1098/rspb.2001.1862.
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 31(1):21–32. doi: 10.1006/fgbi.2000.1228.
- Thiers HD. 1958. The agaric flora of Texas. II. New taxa of white- and pink-spored agarics. Mycologia. 50(4):514–523. doi: 10.1080/00275514.1958.12024747.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180. doi: 10.1111/j.1096-0031.2010.00329.x.
- Vellinga EC, Noordeloos ME. 2001. Glossary. In: Noordeloos M, Kuyper T Vellinga E, editors Flora agaricina neerlandica 5. Lisse/Abingdon/Exton (PA)/Tokyo: A.A. Balkema Publishers; pp. 6–11.
- Vizzini A, Consiglio G, Marchetti M. 2019. Mythicomycetaceae fam. nov. (Agaricineae, Agaricales) for accommodating the genera mythicomyces and Stagnicola, and Simocybe Parvispora reconsidered. Fungal Syst Evol. 3(1):225–240. doi: 10.3114/fuse.2019.03.05.
- Wang G, Yang Z. 2023. Tricholomopsis muconata, a new species from southwestern China. J Fungus Res. 21:24–30.
- Wang JF, Zhou XY, Pan K, Jiang AM. 2018. Tricholomopsis flammula, a new record of the genus Tricholomopsis in China. J Henan Agric Univ. 52(4):621–624. doi: 10.16445/j.cnki.1000-2340.2018.04.022.
- Yamamoto N, Suzuki T, Kobayashi M, Dohra H, Sasaki Y, Hirai H, Yokoyama K, Kawagishi H, Yano K. 2014. A-WINGS: an integrated genome database for Pleurocybella porrigens (Angel’s wing oyster mushroom, Sugihiratake). BMC Res Notes. 7(1):866. doi: 10.1186/1756-0500-7-866.
- Yang ZL, XX D, Kost G, Rexer KH. 2017. New species in the Tricholoma pardinum complex from Eastern Himalaya. Phytotaxa. 305(1):1. doi: 10.11646/phytotaxa.305.1.1.
- Zang M, Zeng XL. 1978. A preliminary study on the family paxillaceae of Yunnan and Tibet, China. Acta Mycol Sin. 18(4):279–364. doi: 10.13343/j.cnki.wsxb.1978.04.001.
- Zdobnov EM, Kuznetsov D, Tegenfeldt F, Manni M, Berkeley M, Kriventseva EV. 2021. OrthoDB in 2020: evolutionary and functional annotations of orthologs. Nucleic Acids Res. 49(D1):D389–D393. doi: 10.1093/nar/gkaa1009.
- Zeng C, Hollingsworth PM, Jing Y, He Z, Zhang Z, Li D, JunBo Y. 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 14(1):43. doi: 10.1186/s13007-018-0300-0.
- Zhao R, Li G, Sánchez-Ramírez S, Stata M, Yang Z, Wu G, Dai Y, He S, Cui B, Zhou J, et al. 2017. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 84(1): 43–74. doi: 10.1007/s13225-017-0381-5.

