ABSTRACT
Ebola virus (EBOV), belonging to the species Zaire ebolavirus in the genus Ebolavirus, causes a severe febrile illness in humans with case fatality rates (CFRs) up to 90%. While there have been six virus species classified, which each have a single type virus in the genus Ebolavirus, CFRs of ebolavirus infections vary among viruses belonging to each distinct species. In this review, we aim to define the ebolavirus species-specific virulence on the basis of currently available laboratory and experimental findings. In addition, this review will also cover the variant-specific virulence of EBOV by referring to the unique biological and pathogenic characteristics of EBOV variant Makona, a new EBOV variant isolated from the 2013–2016 EBOV disease outbreak in West Africa. A better definition of species-specific and variant-specific virulence of ebolaviruses will facilitate our comprehensive knowledge on genus Ebolavirus biology, leading to the development of therapeutics against well-focused pathogenic mechanisms of each Ebola disease.
Introduction
Ebola virus disease (EVD) is an acute viral zoonotic disease with high case fatality rates (CFRs) reaching as high as 90%. The disease is characterized by sudden onset of high fever, gastrointestinal symptoms including diarrhea and vomiting, respiratory symptoms, rash, conjunctival injection, and hemorrhagic manifestations. Fatal cases terminate in hypovolemic shock and multiorgan failure [Citation1,Citation2]. Ebola virus (EBOV), the causative agent of EVD, is a negative-sense, single-stranded RNA virus in the genus Ebolavirus of the family Filoviridae. In the genus Ebolavirus, there have been six virus species classified that each have a single type virus [Citation1,Citation3]. Among them, EBOV is often referred to as the most virulent ebolavirus, in large part because EBOV has been responsible for the majority of Ebola disease outbreaks thus far, including several epidemics with significantly high CFRs (>70%) [Citation1,Citation4,Citation5]. Aside from the epidemiological aspect, some laboratory and experimental findings also indicate that virulence/pathogenicity of ebolaviruses in humans differs among viruses belonging to each distinct species. In this review, we aim to define the virus species-specific difference on the basis of laboratory and experimental findings, including molecular insights. We first outline current knowledge on EBOV pathogenesis based on three aspects, such as clinical, in vivo, and in vitro studies. We then interpret the distinct virulence profiles among the ebolaviruses belonging to the different species and further discuss the research gaps in our understanding of this species-specific pathogenicity of ebolavirus in humans. Finally, this review deliberates recent findings of virulence of the EBOV-Makona variant, a newly identified EBOV variant from the largest EVD outbreak in West Africa in 2013–2016, which exhibits unique virulence characteristics compared to the known EBOV variants/strains.
Molecular biology of ebolaviruses
The EBOV comprises negative-sense, single-stranded RNA genome that consists of seven genes, encoding the nucleoprotein (NP), virion protein 35 (VP35), VP40, the glycoprotein (GP), VP30, VP24, and the RNA-dependent RNA polymerase (L) [Citation6]. Additionally, soluble GP and small soluble GP are encoded by the GP gene [Citation7–9]. The termini of the genome comprise a 3′ leader and a 5′ trailer that contain replication/transcription promoters and genome packaging signals. The EBOV particles possess a ribonucleoprotein (RNP) complex, consisting of the viral genome RNA encapsidated in the NP, VP35, VP30, VP24, and L, surrounded by a viral matrix protein VP40 and a host-derived envelope studded with GP spikes (). The L, along with VP35, a polymerase cofactor, and viral transcription factor VP30 drive replication of the viral genome and transcription of the genes [Citation10,Citation11]. While VP40 is essential for virion assembly and budding [Citation12–14], GP mediates viral entry, including attachment to receptor molecules and membrane fusion [Citation15,Citation16]. The VP24 plays a key role in condensing viral nucleocapsids, which is important for efficient packaging of genome/nucleocapsid into the virion [Citation17–22].
Figure 1. Genomic organization and viral particle of ebolaviruses. The single-stranded, negative-sense genome consists of a linear RNA molecule of approximately 19 kb that is composed of seven genes: NP, VP35, VP40, GP, VP30, VP24, and L. The viral particles are filamentous in shape, consisting of a nucleocapsid core surrounded by a viral matrix protein VP40 and a host-derived envelope studded with GP spikes
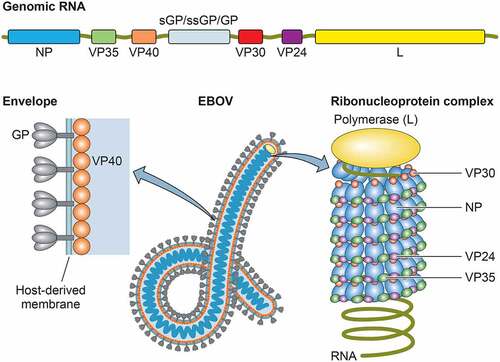
Taxonomy of ebolaviruses
Ebolaviruses belong to the family Filoviridae, which comprises six virus genera, including Ebolavirus, Marburgvirus, Cuevavirus, Dianlovirus, Striavirus, and Thamnovirus [Citation3]. In the genus Ebolavirus, six species that each have a single type virus have been identified to date: Zaire ebolavirus with type virus Ebola virus (EBOV), Sudan ebolavirus with type virus Sudan virus (SUDV), Bundibugyo ebolavirus with type virus Bundibugyo virus (BDBV), Tai Forest ebolavirus with type virus Taï Forest virus (TAFV), Reston ebolavirus with type virus Reston virus (RESTV), and Bombali ebolavirus with type virus Bombali virus (BOMV) [Citation1,Citation3]. The average amino acid identity among all viral proteins between viruses belonging to genus Ebolavirus ranges from 60% to 80% [Citation23].
Note that the term “species” used throughout this review refers to “virus species.” In addition, we use the term “Ebola disease” to describe all diseases caused by the infection of viruses belonging to genus Ebolavirus, and the terms “Ebola virus disease (EVD),” “Sudan virus disease (SVD),” and “Bundibugyo virus disease (BVD)” to describe each Ebola disease that is subcategorized according to its causative agent [Citation24].
Outbreaks and CFRs for ebolavirus infections
Although all ebolaviruses, except for BOMV, are known to infect humans, CFRs of ebolavirus infections vary, ranging from no fatalities (0% CFR) in RESTV and TAFV infections to reaching up to 90% of EBOV infection () [Citation5]. EBOV has been responsible for the majority of Ebola disease outbreaks to date since its first discovery in 1976, including the largest EVD outbreak in history that occurred in West Africa in 2013–2016 [Citation1,Citation4,Citation5]. Despite its magnitude, the CFR for the 2013–2016 West Africa EVD outbreak was 40%, which was exceptionally low compared to other EVD outbreaks reported so far. Multiple factors, including a better international outbreak response, such as robust clinical trials and clinical interventions, likely contributed to the observed low CFRs in the West Africa EVD outbreak. A possible involvement of a virological factor in the low CFRs will be discussed in Section “Pathogenesis of EBOV-Makona variant”. The average CFR for EVD excluding the 2013–2016 EVD outbreak is 72% ().
Figure 2. Ebola disease outbreaks and case fatality rates (CFRs) of EBOV, SUDV, and BDBV infections. Forest plot shows the average CFRs and 95% confidence intervals (CIs) for the Ebola disease outbreaks by fixed-effect model with inverse-variance weighting. Meta-analysis was performed using R software with the metaphor package. The CFRs from the 2013–2016 EVD outbreak caused by EBOV-Makona variant is separately calculated from the average CFRs from all other EVD cases. N.A: not available due to small sample size
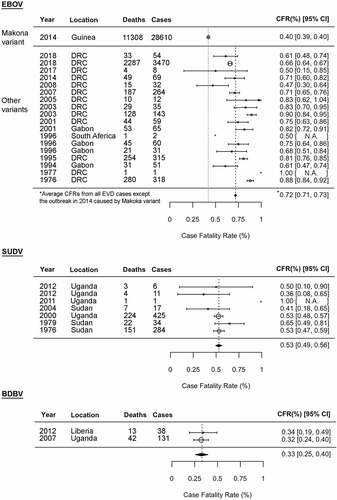
SUDV was first discovered in 1976, and thus far has been responsible for seven outbreaks [Citation25,Citation26]. The largest SVD outbreak in 2000 resulted in more than 400 human infections with a CFR of 53%. Compared to EBOV and SUDV, BDBV is often referred to as a less virulent ebolavirus. Two BVD outbreaks have been reported in 2007 [Citation27] and 2012 [Citation28–30], with culminating average CFR as 33%. To date, there has been only one confirmed case of severe, but nonfatal, TAFV infection in humans reported in 1994 [Citation31,Citation32]. A total of 74 persons were defined as direct contact persons with the patient, but all were shown as seronegative for ebolaviruses [Citation32].
While EBOV, SUDV, BDBV, and TAFV cause often severe/fatal diseases in humans, RESTV is apparently apathogenic to humans. There has been no evidence showing that RESTV is associated with human disease since its first discovery in 1989 [Citation33], despite the apparent occurrence of human infections evidenced by seropositive titers of RESTV-specific antibody that relates to several RESTV epizootics in nonhuman primates (NHPs) or domestic pigs [Citation34–37].
BOMV was first discovered in bats in 2018 [Citation38], followed by the second identification in bats in 2019 [Citation39]. The potential of this virus to infect/cause disease in humans is currently unknown.
Pathogenesis of EVD and its molecular mechanisms
Studies on ebolavirus pathogenesis have mostly been focused on EBOV and its disease. Owing to numerous studies performed during the 2013–2016 West Africa EVD outbreak, detailed clinical insights on EVD have also become available. In this section, we will provide current knowledge on EVD pathogenesis at both in vivo and molecular levels. Note that we will mainly focus on the aspects of which insights are also available in studies of other ebolavirus species. The comparison between EVD (EBOV) and other Ebola diseases (other ebolaviruses) will be provided in Section “Comparison of pathogenesis among viruses belonging to genus Ebolavirus”.
Clinical and laboratory findings on EVD
The initial symptoms of EVD are a nonspecific febrile illness followed by gastrointestinal manifestations [Citation1,Citation2,Citation40–50]. Some respiratory symptoms, hiccup, conjunctival injection, and macropapular rash are also often reported. Hemorrhagic signs are observed in approximately half of the infected persons, with no clear correlation between bleeding and disease severity [Citation44,Citation51,Citation52]. The mean incubation period for EVD from contact exposure to the onset of symptoms is 7.34 ± 1.35 d [Citation53]. In fatal cases, death occurs 6–16 d after the onset of symptoms as a result of hypovolemic shock and multiorgan failure [Citation2].
Acute, robust, and systemic viral replication is the most consistent observation in severe/fatal EVD. Viral antigen and nucleic acid can be detected in patients’ blood from d 1 to 3, peaking at around 6–7 d post symptom onset, and remain high throughout the course of the disease in fatal cases [Citation1,Citation42,Citation54,Citation55]. A number of clinical studies have shown a strong correlation between viremia titer and EVD fatality [Citation40–42,Citation44,Citation50,Citation55–57]; high blood viral load (≥106 copies/ml) was found to be predictive of the fatal outcome [Citation41,Citation44]. Markedly elevated levels of alanine aminotransferase (ALT), which is a biochemical marker for liver integrity, and aspartate aminotransferase (AST), which is a biochemical marker for liver, heart skeletal muscle, kidney, brain, and red blood cell integrity, are major serum biochemical signatures seen in severe/fatal EVD, indicating serious tissue damage due to EBOV infection [Citation44].
Over-activated, detrimental pro-inflammatory cytokine/chemokine responses, such as the so-called cytokine/chemokine storm, are also one of the most common patho-immunological features in severe/fatal EVD [Citation42,Citation55,Citation57–65]. Massive production of multiple pro-inflammatory cytokines [e.g. interleukin 1 beta (IL-1β), IL-6, tumor necrosis factor alpha (TNF-α)] and pro-inflammatory chemokines [e.g. IL-8, macrophage inflammatory protein 1 alpha (MIP-1α), MIP-1β, monocyte chemotactic protein 1 (MCP-1), macrophage colony-stimulating factor (M-CSF), interferon gamma-induced protein 10 (IP-10)] are detected in deceased EVD patients. This over-activation of pro-inflammatory mediators results in systemic inflammatory response syndrome (SIRS) that also triggers mixed/compensatory anti-inflammatory response syndrome (MARS/CARS). Indeed, up-regulation of anti-inflammatory cytokines (e.g. IL-1RA, IL-10, sTNF-RI, sTNF-RII) are also observed in EVD patients. This aberrant immunological status, such as SIRS coupled with MARS/CARS, is characteristic of classical bacteria sepsis [Citation66] and has been known to play a significant role in induction of endothelial dysfunction, vascular leakage, and coagulation abnormalities.
Uncontrolled pro- and anti-inflammatory cytokine responses are also linked to the impairment of host adaptive immunity seen in fatal EVD patients. Ruibal et al. have demonstrated that, compared to survivors, EVD fatalities had a higher percentage of T cells expressing inhibitory molecules CTLA-4 and PD-1, despite no difference in the activation status of T cells between fatalities and survivors [Citation65]. Moreover, while strong EBOV-specific T cell responses were detected in EVD survivors [Citation67–69], antigen-specific T cell responses were found to be very rare in fatal cases [Citation65], indicating nonspecific, dysfunctional T cell activation in EVD fatalities. Accordingly, the humoral response mediated by B cells was also impaired in fatal EVD cases [Citation55,Citation61,Citation70]. In sum, robust viral replication and immune dysfunction are key hallmarks in severe/fatal EVD patients, significantly contributing to EVD pathogenesis.
EBOV pathogenesis studied in animal models
Animal models of EBOV infection have so far been developed in NHPs and small animal models, such as mice, guinea pigs, Syrian golden hamsters, and ferrets. Among them, NHPs are considered the “gold standard” animal model since they are highly susceptible to the infection of wild-type EBOV (WT-EBOV) that have been isolated from human samples. Nearly all clinical and pathological features of severe/fatal EVD in humans can be recapitulated in NHPs, including high viremia, strong cytokine/chemokine response, coagulopathy, rash, and hemorrhagic signs [Citation71–75]. Cynomolgus and rhesus macaques have been the most commonly used NHPs for EBOV infection; disease progression in cynomolgus macaques after EBOV infection is slightly faster than that in rhesus macaques [Citation6]. Pathological studies in EBOV-infected NHPs have clearly demonstrated that cells of the mononuclear phagocytic system (MPS) (i.e. monocytes, macrophages) and dendritic cells (DCs) are the initial target cells [Citation71,Citation73,Citation76] that migrate to target organs (e.g. liver, lymph nodes, spleen), resulting in the efficient transmission and replication of the virus [Citation77]. In addition to NHPs, lethal infection with WT-EBOV can also be achieved in ferrets with hallmark pathological features of EVD, including fever, petechial rashes, hemorrhage, and coagulopathy [Citation78].
Unlike NHPs and ferrets, immunocompetent rodents show no or very mild signs of disease after WT-EBOV infection. Thus, immunocompetent rodent models rely on using rodent-adapted virus strains that have been established as a consequence of serial passage of the virus in the host animals, such as mice or guinea pigs, leading the virus to acquire the ability to cause uniformly lethal infection in rodents [Citation79,Citation80]. Two of the mouse-adapted EBOV strains (MA-EBOV) have been developed based on EBOV variant Mayinga [Citation81] and Makona [Citation82], and four of the guinea pig-adapted EBOV strains (GPA-EBOV) are available [Citation79,Citation80,Citation83–86]. Rodent-adapted EBOV targets the same cells/tissues as the WT-EBOV infection in the lethally susceptible animals (i.e. NHPs, humans) and develops high viremia in the infected host rodents [Citation81,Citation83,Citation87]. MA-EBOV can also cause lethal infection in Syrian golden hamsters with several critical EVD signatures including cytokine/chemokine responses and coagulopathy [Citation88]. A collaborative cross (CC) resource recombinant inbred (RI) intercrossed (CC-RIX) mouse model with MA-EBOV infection has also been developed, which shows severe coagulopathy that is not evident in other conventional laboratory mouse strains [Citation89].
Furthermore, several lethal immunodeficient rodent models with WT-EBOV infection have been developed and widely used for EBOV pathogenesis studies. In addition to the classical immunodeficient mice, such as type I interferon (IFN)-deficient mouse strains (i.e. IFN-α/β receptor knockout (IFNAR−/-) mice [Citation90,Citation91], STAT1 knockout (STAT1−/-) mice [Citation92]), and adaptive immunity-deficient mouse strain (i.e. SCID mice) [Citation90], several human immune system (HIS) mice, generated by xeno-engrafting human immune cells or tissues and/or their progenitors into immunodeficient mice, have also been utilized for studies of EBOV pathogenesis [Citation93–100]. The use of HIS mice allows for the examination of the cell/organ-specific human immune response and its interaction with virus in vivo, thus they are considered a valuable tool for studying EVD pathogenesis.
Molecular pathogenic mechanisms of EBOV infection
Among all EBOV proteins, VP35, VP24, and GP have been considered as the main virulence factors of EBOV. VP35 functions as an IFN antagonist, which inhibits host type I IFN-α/β induction, and also inhibits the phosphorylation of double-stranded RNA-activated protein kinase R that mediates cellular antiviral responses [Citation101–110]. The introduction of mutations disabling the VP35’s IFN-antagonistic function results in attenuation of virulence in animal models including NHPs [Citation103,Citation108,Citation111–113]. While VP35 counteracts the type I IFN induction pathway, VP24, the other EBOV IFN antagonist, blocks the type I IFN signaling cascade by blocking karyopherin α (KPNA)-mediated nuclear translocalization of phosphorylated STAT1 homodimers or STAT1-STAT2 heterodimers [Citation114–117]. The restriction of STAT nuclear translocalization by VP24 results in reduced transcriptional activation of IFN-stimulated genes (ISGs), preventing the establishment of an antiviral state in the host cells. The significant role of VP24 for the acquisition of virulence in rodents has been reported [Citation79,Citation84–86,Citation118–120]. VP35 and VP24 also contribute to suppression of DC activation/maturation, resulting in induction of impaired cell-mediated responses [Citation121–124], as also seen in EBOV-infected DCs [Citation125,Citation126].
The GP has been shown to interact with host toll-like receptor 4 (TLR4) leading to the activation of pro-inflammatory responses via the NF-κB pathway [Citation127–131]. In addition, the shed GP, which is a truncated version of the surface GP cleaved by cellular metalloprotease TACE, has also been shown to induce immune activation in a TLR4-dependent manner, aside from its antibody-neutralizing activity and endothelial-permeabilizing activity [Citation132–134]. Moreover, a recent study demonstrated the interaction of virion-associated phosphatidylserine with Tim-1 and showed its significant role in T-cell activation, production of pro-inflammatory mediators, and pathogenesis in the animal model [Citation135].
Comparison of pathogenesis among viruses belonging to genus Ebolavirus
The CFRs of Ebola diseases in humans can range from no fatalities (0% CFR) in RESTV and TAFV infections to reaching up to 90% of EBOV infection (). Although clinical insights on the SUDV, BDBV, TAFV, and RESTV infections in humans are limited, there have been intriguing experimental in vivo and in vitro studies available that examine differing pathogenesis of these viruses. Several small animal models (i.e. ferrets, humanized mice) that propagate lethal SUDV, BDBV, TAFV, and RESTV infections have also been developed, greatly facilitating the studies on ebolavirus pathogenesis. In this section, we will compare the pathogenesis of EVD (discussed in Section “Pathogenesis of EVD and its molecular mechanisms”) with other Ebola diseases and define species-specific differences on the basis of experimental findings ( and ).
Figure 3. Species-specific and variant-specific virulence of ebolaviruses. Viral ability to spread from macrophages to parenchymal cells, leading to severe organ damage, is one of the key phenotypic features determining ebolavirus pathogenicity. The numbers 1–6 shown in a table on the right are correlated with those shown in a figure on the left. *Including Kupffer cells. na: not available
Disease courses and fatalities in animal models
Differing disease severity and lethality associated with each ebolavirus infection have been demonstrated by several animal studies (). In the case of NHP models, EBOV causes 100% lethality in cynomolgus macaques by 5–7 d [Citation136–139] and in rhesus macaques by 6–9 d [Citation74,Citation136,Citation140] after infection with a challenge dose of 103 PFU via intramuscular route. In contrast, SUDV, BDBV, TAFV, and RESTV challenges do not cause uniform lethality in cynomolgus macaques, and the median time to death in macaques infected these viruses appears to be longer than EBOV infection; SUDV causes 50–100% lethality in 9–10 d after infection with a dose of 103 PFU [Citation6,Citation137,Citation139], BDBV causes 50–75% lethality in 10–13 d after infection with a dose of 103 −104 TCID50 or 103 PFU [Citation141–143], TAFV causes 60% lethality in 10–14 d after infection with a dose of >103 PFU [Citation139,Citation144], and RESTV causes 80–100% lethality in 8–21 d after infection with a dose of 103 PFU [Citation6,Citation145]. Considering the fact that a lethal infection in macaques can be achieved by EBOV infection with challenge doses lower than 103 PFU – as low as 0.01 PFU or as little as one infectious unit – with various inoculation routes [Citation146–149], SUDV, BDBV, TAFV, and RESTV seem to be inherently less virulent than EBOV in macaques. Parenthetically, RESTV seems to be a quite unique virus among ebolaviruses and is presumably an animal pathogen given its distinct virulence in macaques and humans. In addition to NHP studies, a longer duration of disease after experimental infection of SUDV and BDBV compared to EBOV has also been reported in a ferret model [Citation78,Citation150,Citation151].
Table 1. Comparison of fatality rates and time to deaths in animal models infected with different ebolaviruses
Table 2. Comparison of pathogenic functions of viral proteins among ebolaviruses
Notably, recently developed humanized mouse model, HLA-A2-transgenic NOD-scid-IL2γ receptor-knockout mice reconstituted with human hematopoiesis (huNSG-A2), showed a distinct susceptibility to ebolaviruses belonging to each distinct species, and the infection recapitulated species-specific CFRs in humans; EBOV, SUDV, BDBV, TAFV, and RESTV infections caused lethality of 92.3%, 71.4%, 28.6%, 18.2%, and 20% in mice, respectively [Citation98]. This suggests that HIS greatly contributes to ebolavirus species-specific pathogenesis.
Replication ability
Similarly to EVD, a correlation of high viremia titer and fatal disease outcome is commonly observed in SUDV disease (SVD) patients [Citation52] and BDBV disease (BVD) patients [Citation152]. However, very limited but some pathological examinations of postmortem specimens of fatal EVD, SVD, and BVD patients have shown that replication of SUDV and BDBV in human cells/organs might be less efficient than EBOV [Citation54,Citation153,Citation154]. A previous comparative histopathological analysis showed that, whereas extensive distribution of EBOV antigen and virus inclusions composed of aggregates of viral nucleocapsids was detected in liver from EVD fatalities, no virus inclusions and the least amount of virus antigen distribution were found in liver from BVD fatalities, and SUDV infection was somewhat intermediate () [Citation54]. Notably, the level of ALT, a biochemical marker for hepatocyte integrity, in fatal SVD cases was below 100 U/L [Citation155], which is significantly lower than that generally seen in fatal EVD cases with around 500 U/L (often more than 1000 U/L) [Citation42,Citation156]. Moreover, while disease severity is often correlated with blood ALT levels in the EVD patients, comparable levels of ALT were observed between fatal and nonfatal cases of SVD [Citation155]. These clinical insights suggest that abilities for replication and/or causing tissue damage in liver might differ between EBOV and other ebolaviruses.
Consistent with lower levels of replication found in postmortem specimens of fatal BVD, a reduced replication ability of BDBV was also found in an in vitro study using human peripheral blood mononuclear cells (PBMCs). The study showed that, whereas EBOV titer in supernatants of infected PBMCs peaked in 5 d, BDBV titer peaked at 8 d postinfection with a titer approximately 1-log lower than that of EBOV [Citation157]. Interestingly, electron microscopic analysis showed the accumulated nucleocapsids in macrophages infected with BDBV, suggesting that BDBV can infect and replicate in human macrophages but cannot be released as efficiently as EBOV ().
A slower and lower growth ability in vitro was also found in apathogenic RESTV compared to EBOV using Vero or Vero E6 cells [Citation158,Citation159]. Moreover, several studies using an ebolavirus minigenome system, which allow to model ebolavirus replication/transcription without using infectious virus in biosafety level 4, have suggested that the polymerase complex of RESTV exhibited a lower ability of viral RNA synthesis than that of EBOV () [Citation158,Citation160]. Interestingly, supplying of EBOV RNP complex proteins, such as NP, VP35, VP30, and L, in the RESTV minigenome assay system produced a higher reporter transcription activity than using RESTV RNP complex [Citation160]. Although it has been considered that ebolaviruses share a common strategy for their genome transcription/replication, thus the same viral proteins are required for these processes, the performance of the RNP complex for driving efficient genome transcription/replication might vary among ebolaviruses.
The intriguing difference between pathogenic and apathogenic ebolaviruses can be found in their distinct replication characteristics in parenchymal cells of the liver, which the primary target organ of EBOV replication in vivo. The immunohistochemistry analysis using immunodeficient mouse models demonstrated the wide distribution of EBOV antigen in the liver, by infection of hepatocytes, endothelial cells, and Kupffer cells, which are the first target cells of the liver. This is in stark contrast to RESTV replication, which has been noted to be very limited in the liver and mostly restricted to Kupffer cells () [Citation96,Citation98,Citation159]. Consistent with this, a few in vitro studies have shown that RESTV replicates in primary human macrophages [Citation130] and human monocytic leukemia THP-1 cells [Citation161] similarly to EBOV, but not as efficient as EBOV in human hepatocellular carcinoma Huh7 cells at early phase of infection [Citation162]. Interestingly, this distinct replication pattern in liver between pathogenic EBOV and apathogenic RESTV is also observed in a comparative study between lethal MA-EBOV and non-lethal WT-EBOV infections in Syrian golden hamsters [Citation88]. These findings strongly suggest that the ability of virus to spread from macrophages to parenchymal cells is a key signature of pathogenic ebolaviruses.
The efficient viral replication in liver is not only responsible for its systemic spread but also the cause of significant hepatocyte necrosis manifested as severe liver damage in EVD fatalities and EBOV-infected animals. Importantly, dysfunction of the liver contributes to coagulation disorders – one of the key pathological features in EVD – due to impairment in the synthesis of important clotting factors. The correlation of viral replication in the liver and disease severity in Ebola disease pathogenesis has been widely acknowledged; however, experimental evidence to precisely compare ebolavirus replication ability in human hepatic cells is not currently available. Thus, it remains unclear whether the less efficient replication of less virulent/apathogenic ebolaviruses in liver is due to their respectively poorer inherent abilities to infect or replicate in hepatocytes or due to an unknown host antiviral mechanism(s) that interferes with viral spread from macrophages to hepatocytes. Basic characterization of viral replication ability in hepatocytes (e.g. human primary hepatic cells) is necessary to understand species-specific pathogenesis of ebolaviruses.
Induction of uncontrolled pro-inflammatory responses
The activation of pro-inflammatory responses is considered as a common feature of fatal Ebola diseases;however, the types of cytokines/chemokines upregulated by the infection seem to be varied among viruses belonging to each distinct species. For example, the expression levels of TNF-α and IFN-γ, which have been found to be increased in EVD fatalities, were shown to be low in the SUDV infection in humans [Citation163]. Moreover, a previous clinical study that compared cytokine expression profiles between samples from acute phase (collected within 11 d from the onset of illness) received from both BVD fatalities and survivors, and convalescent phase from BVD survivors, showed significantly lower IL-1α, IL-1β, IL-6, and TNF-α levels in acute samples than in convalescent samples () [Citation152]. The levels of these cytokines expressed in BVD patients were even lower than those in the healthy control group, suggesting that unlike EBOV, BDBV infection might down-regulate these cytokine expressions in humans. A distinctive pattern of cytokines/chemokines expression induced by BDBV infection was also found in in vitro study [Citation157]. BDBV infection in human PBMCs induced the expression of IL-1β, MIP-1α, TNF-α, and MCP-1 at significantly lower levels than EBOV infection. In particular, very low level of IL-1β (30–70 pg/mL) was produced in BDBV-infected PBMCs, in contrast to the EBOV infection that induced IL-1β with a peak concentration >2500 pg/mL. Intriguingly, fever with above 38°C was less common in the 2012 BVD outbreak; only 38.9% of BVD patients showed high fever during hospital stay [Citation29]. This atypical clinical manifestation of BVD might be related to the distinct characteristic of BDBV for the regulation of pro-inflammatory responses.
A previous in vitro study using virus-like particles containing RESTV GP demonstrated that, unlike EBOV GP, RESTV GP does not trigger TLR4 signaling () [Citation130]. In agreement with this, a significantly weaker cellular response was observed in human primary monocytes or monocyte-derived macrophages (MDMs) infected with RESTV, in contrast to the strong activation of MDMs by EBOV infection shown as a markedly elevated level of pro-inflammatory genes [Citation130,Citation164]. These results suggest that macrophage activation mediated by GP via interaction with TLR4 might be a critical factor that determines the ebolavirus pathogenicity. On the other hand, while TLR4 inhibitors showed efficacy in the EBOV mouse model, TLR4 knockout mice still exhibited full susceptibility to the lethal MA-EBOV infection [Citation165]. This suggests that there might be an additional factor(s), which critically involves in detrimental pro-inflammatory responses in EVD. Further analysis will be needed in order to not only precisely characterize pro-inflammatory activation profiles for each ebolavirus infection, but also define the molecular mechanisms underlying uncontrolled pro-inflammatory responses seen in Ebola diseases.
Disturbance of functional immune responses
IFN antagonisms mediated by EBOV VP35 and VP24 have been well-characterized by a number of in vitro studies as described in Section “Molecular pathogenic mechanisms of EBOV infection”. Thus, the function of these viral proteins from other ebolaviruses has been closely studied in order to find a clue that determines differing pathogenicity among ebolavirus species. However, the apparent difference in terms of the type I IFN-antagonism between EBOV and other ebolaviruses has not been well-defined. It was shown that VP35 proteins from SUDV, BDBV, TAFV [Citation166,Citation167], and RESTV [Citation102,Citation166–169] are all able to inhibit type I IFN induction in the same manner as EBOV VP35 (). Moreover, whereas WT-EBOV infection becomes lethal in IFNAR−/- mice or in mice treated with antibodies against IFN-α/β, the BDBV, TAFV, and RESTV infections still fail to cause lethal illness in IFNAR−/- mice [Citation90,Citation91]. These observations suggest that, although the inhibition of the host type I IFN production – presumably solely mediated by VP35 – is critical for ebolaviruses to successfully and efficiently initiate replication in the host, this function is not the main virulence determinant among ebolavirus species.
On the other hand, species-specific difference might exist in their abilities for inhibition of type I IFN-signaling pathway as well as ISGs expression. Schwards et al. demonstrated a lower ability of BDBV VP24 to bind KPNA (KPNA1, KPNA5, and KPNA6) compared to EBOV VP24, consistent with a decreased inhibitory effect of BDBV VP24 on the expression of ISGs [Citation170] that might explain the lower virulence of BDBV than EBOV. Moreover, a previous in vitro study using infectious EBOV and RESTV demonstrated that, whereas the expression of a number of ISGs was down-regulated in IFN-treated Huh7 cells with EBOV infection, RESTV infection failed to down-regulate those ISG expression [Citation162]. This suggests that RESTV has a diminished capacity to counteract host IFN-signaling; however, this mechanism cannot be explained by its VP24’s function; although some amino acid differences were found between RESTV VP24 and EBOV VP24 [Citation171–173], RESTV VP24 has an intact KPNA-binding capability or ISG inhibition () [Citation170]. Furthermore, it was shown that STAT1−/- mice infected with RESTV survived with disease signs appearing slower and milder than those infected with EBOV [Citation92,Citation174], suggesting that differing pathogenicity between EBOV and RESTV cannot be solely explained by mechanism(s) underlying inhibition of the Jak/STAT-signaling pathway. Aside from the IFN-antagonistic functions of VP35 and VP24, their abilities to block DC maturation will be interesting to compare among ebolaviruses, which may shed the light on the species-specific immune disturbance.
A possible species-specific difference might exist in host adaptive responses against ebolavirus infections; while elevated IgG levels in non-survivors were rarely seen during the disease course of EVD [Citation55,Citation61,Citation70], high IgG titers were observed in most of the BVD patients including nonsurvivors, corresponding to high virus antigen titers [Citation152]. This finding suggests that BDBV infection may induce stronger humoral immunity than EBOV. In addition, Nehls et al. recently reported that secretory vesicles containing EBOV GP, so-called EBOV GP-virosomes, have the ability to capture neutralizing antibodies and also showed that the efficiency of the virosome formation of GPs from other ebolaviruses was lower than EBOV GP [Citation175]; however, the practical contribution of this mechanism to viral pathogenesis has remained elusive. More experimental evidence will be needed to understand the species-specific difference in host adaptive immunity.
Pathogenesis of EBOV-Makona variant
As discussed above, ebolavirus species-specific distinctions in pathogenicity in humans is one of the intriguing unsolved research topics in the filovirus field. Besides, the virulence of novel EBOV variant Makona, which was responsible for the 2013–2016 West Africa EVD outbreak, the largest EVD outbreak ever documented, is an emerging compelling question to be addressed for a better understanding of the EVD outbreak and EBOV pathogenesis. In this section, we discuss the biological and pathogenic differences between the EBOV-Makona variant and previously identified EBOV variants in humans based on current findings.
Did EBOV-Makona variant acquire enhanced virulence during the outbreak?
EBOV-Makona variant was responsible for the 2013–2016 EVD outbreak that widely spread from Guinea to other countries in Western Africa, ending up a total of >28,600 human infections and >11,300 deaths [Citation5]. During the outbreak, the number of human infections increased day-by-day with unprecedented scale, which had given rise to fears that the virus adapted to humans due to extensive human-to-human transmission events, resulting in the acquisition of enhanced transmissibility and/or virulence in humans. The discussion on the basis of sequencing analyses using available EBOV-Makona isolates was therefore primarily focused on (1) whether the EBOV-Makona is mutating more rapidly than usual and (2) whether there is any mutation(s) in the EBOV-Makona genome that possibly plays a significant role in host specificity, viral fitness, transmissibility, or virulence. Various genome analyses of more than 1500 full-length EBOV-Makona sequences – approximately 5% of those infected – have confirmed the accumulation of numerous mutations in later isolates compared to the first isolate from Guinea [Citation176]. However, the mutation rate, which is expressed as the number of mutations per site per year, of the EBOV-Makona during this outbreak was shown to be similar to the previous outbreaks [Citation177–181].
The large-scale sequence data clearly showed the presence of two major lineages; the first one includes isolates sampled from Guinea during the early phase of the epidemic, and the second one contains a majority of isolates sampled from all of the affected countries during the later phase of the epidemic [Citation176,Citation182,Citation183]. Among all nonsynonymous mutations discovered in the viral genome of EBOV-Makona isolates in the second lineage, an amino acid substitution (Ala to Val) on GP at residue 82 (GP-A82V) was most intensively studied for its possible involvement in viral pathogenic characteristics. This was due to the emergence of the A82V mutation at the branch point that distinguishes two lineages and was fixed in the population afterward [Citation182–186], and residue 82 on GP is located at the receptor-binding interface [Citation187]. Several in vitro studies using EBOV GP-pseudotyped virus vectors or EBOV VLP systems have demonstrated that GP-A82V promotes viral entry into human cells [Citation182,Citation183,Citation188], by which the mechanisms were further identified as cathepsin B and Niemann–Pick C1 dependent [Citation189,Citation190]. Interestingly, the enhancement of viral entry mediated by GP-A82V was only observed in primate cells, but not in rodent, carnivore [Citation182], or bat-origin cells [Citation183,Citation191], suggesting that the A82V mutation might have contributed to elevated EBOV-Makona fitness in human population.
Although the above findings also raised a hypothesis in which the virulence of EBOV-Makona in humans was enhanced due to the acquisition of GP-A82V, this hypothesis has been challenged by several in vivo studies. Marzi et al. infected rhesus macaques with EBOV-Makona early isolates containing GP-82A and EBOV-Makona later isolates containing GP-82V and found no statistically significant difference in survival rate, tissue/blood viral load, IFN response, serum biocheminal parameters, or coagulation parameters among the infected groups [Citation192]. Interestingly, the animals infected with EBOV-Makona later isolates showed even relatively prolonged disease progression and reduced tissue/blood viral load compared to those infected with EBOV-Makona early isolates. Moreover, in vitro work indicated that later isolates of EBOV-Makona replicated in human hepatocyte Huh-7 cells less efficiently than earlier isolates [Citation192], which was contrary to the idea that the GP-A82V mutation enhances the EBOV-Makona infectivity in human cells. These results obtained by using infectious virus strongly suggest that, although it is still possible that the GP-A82V mutation has changed the biological characteristics of EBOV-Makona to some extent, those effects were most likely canceled by other mutations in GP or other viral genes in EBOV-Makona later isolates.
A mutation that is possibly involved in the attenuated phenotype of the EBOV-Makona later isolates is an amino acid substitution (Asp to Gly) on L viral RNA polymerase at residue 759 (L-D759G) [Citation191]. Indeed, while 40% of IFNAR−/- mice died after the infection of EBOV-Makona early isolate containing 759D in L, none of the mice died by the infection of a recombinant EBOV-Makona mutant in which the amino acid position 759 on L is substituted from asparagine to glycine. Moreover, a significantly delayed time to death was observed in ferrets infected with L-D759G mutant compared to WT virus. Virulence evolution, in association with the acquisition of mutation(s), is one of the compelling topics in the field of infectious diseases that often attracts a lot of attention. However, this needs to be carefully and accurately interpreted based on the combination of genomics/phylogenomics studies and confirmatory laboratory-based studies using infectious viruses.
Variant-specific virulence between Makona variant and previously identified variants
It is important to note that several in vivo studies using animal models have strongly suggested that the EBOV-Makona variant, regardless of its isolates, is inherently less virulent than historical EBOV variants that were responsible for previous outbreaks [Citation138,Citation193–196]. Remarkably, whereas the infection of EBOV-Mayinga, the prototype EBOV variant isolated from the 1976 EVD outbreak, resulted in 100% lethality in IFNAR−/- mice as has been known, none of the infected mice succumbed to the infection of either EBOV-Makona early isolate or later isolate [Citation192]. In addition, a delayed disease progression was observed in EBOV-Makona-infected rhesus macaques and cynomolgus macaques compared to the infection of EBOV-Mayinga variant [Citation138,Citation192] or EBOV-Kikwit variant, isolated from the 1995 EVD outbreak [Citation194,Citation195]. Of note, Jankeel et al. have recently reported that viral replication, histopathological changes (e.g. liver necrosis), and inflammation in target organs were less severe in EBOV-Makona-infected macaques compared to EBOV-Kikwit-infected macaques [Citation196]. In the case of human infections, while the EBOV-Makona caused the largest EVD outbreak in history, the overall CFR during this epidemic was 40%, which was significantly lower than CFRs of previous EVDs caused by EBOV-Mayinga or -Kikwit (CFRs = 88% or 81%, respectively). It is difficult to definitively conclude the virulence of ebolaviruses in humans according to the reported CFRs, which are easily affected by multifactorial causes including not only pathogen-related factors but also social/environmental factors such as medical infrastructures at the outbreak sites/countries or host genetic factors affecting the immune responses against virus infection. However, these virological observations strongly suggest that the EBOV-Makona variant is very unique among previously identified EBOV variants (). While there have been some EBOV variants isolated thus far, variant-specific virulence of EBOV has not been closely investigated. Further studies will be needed to more clearly define biological and pathogenic differences among EBOV variants.
Conclusions and future directions
This review outlined some critical abilities of EBOV for causing severe disease in humans, including robust replication and induction of dysfunctional immune responses (). However, virulence of other ebolaviruses should not also be underestimated. For example, although human infection with TAFV has caused only one nonfatal case, the patient showed severe Ebola disease signs (e.g. diarrhea, vomiting), central nervous system disorder, coagulopathy signs including macular rash and thrombocytopenia, and drastic weight loss – 10% of the initial weight – in 15 d [Citation32]. Moreover, the evidence showing 20% of huNSG-A2 mice succumbing to RESTV infection [Citation98] might suggest that RESTV may have the potential to cause disease in humans under specific conditions (e.g. immune suppression).
While this review highlighted intriguing experimental findings in differing characteristics of ebolaviruses, there are still some significant gaps in our knowledge about ebolavirus species-specific distinctions in pathogenicity at the molecular level. This is in part because, compared to EBOV, our understanding of the basic biology of SUDV, BDBV, TAFV, and RESTV has largely lagged behind. For example, although viral replication in the liver has been known to be critically involved in the pathogenesis of Ebola disease, replication of SUDV, BDBV, TAFV, and RESTV in hepatocytes (e.g. human primary hepatic cells) has not been fully characterized yet. Head-to-head comparison of replication abilities of EBOV with other ebolaviruses in various cell types will provide fundamental information on ebolavirus biology, which is also necessary to evaluate the pathogenic characteristics of each ebolavirus. The application of virus life cycle modeling systems, such as minigenome and transcription/replication-competent VLP systems, would also provide valuable insights into the molecular mechanisms of differing replication characteristics among ebolaviruses.
In vivo analysis using recombinant chimeric ebolaviruses between pathogenic EBOV and apathogenic/less virulent ebolaviruses generated by reverse genetics will also facilitate the identification of ebolavirus virulence determinant(s). Interestingly, a chimeric EBOV possessing RESTV GP replicated in macrophages similarly to the parental EBOV but decreased infection of hepatocytes in infected IFNAR−/- mice [Citation159], suggesting that the RESTV GP may have a deficient interaction with hepatocyte-specific ebolavirus receptor(s), which has not been characterized yet. On the other hand, a chimeric RESTV possessing EBOV GP did not restore the ability of the parental RESTV to replicate in hepatocytes, implying that the ability of virus to spread from macrophages to parenchymal cells – a key signature of pathogenic ebolaviruses () – seems to be defined by multifactorial determinants. The rapidly developing single-cell RNA sequencing technology would be one of the most powerful approaches that enable not only precise identification of virus-infected cell type(s) in target organs, but also a comprehensive understanding of differing host immune responses against each ebolavirus infection at the single-cell level.
One of the significant limitations in conducting comparative pathogenesis studies among ebolavirus species was a lack of animal models that recapitulate differing Ebola diseases in humans. Using recently developed HIS mouse models in future comparative studies will facilitate the understanding on the interaction between virus and host immunity in each Ebola disease at the molecular level. In addition, data sets from several large-scale analyses, including specificity determining positions analysis [Citation173], global phosphoproteomic analysis [Citation197], and multi-platform ’omics analysis [Citation58], have become available. Utilizing these comprehensive data sets, in combination with authentic virological approaches including utilization of reverse genetics for the in-depth understanding of molecular virology aspects, may help to define different characteristics among ebolaviruses. Further studies examining species-specific and variant-specific virulence of ebolaviruses at the molecular level will facilitate not only a better understanding of the basic biology of genus Ebolavirus but also the development of therapeutics against well-focused pathogenic mechanisms of each Ebola disease.
Acknowledgments
We thank Brady Zell for helpful comments and editing the manuscript. We also thank Patrick Lane for improving figure quality.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jacob ST, Crozier I, Fischer WA 2nd, et al. Ebola virus disease. nature reviews disease primers. Vol. 6, Springer US; 2020.
- Campion EW, Feldmann H, Sprecher A, et al. Ebola. N Engl J Med. 2020. DOI:10.1056/NEJMra1901594
- Kuhn JH, Amarasinghe GK, Basler CF, et al. ICTV virus taxonomy profile: filoviridae. J Gen Virol. 2019;100(6):911–912.
- Mylne A, et al. A comprehensive database of the geographic spread of past human ebola outbreaks. Sci Data. 2014;1(1):1–10.
- Centers for Disease Control. History of ebola virus disease. https://www.cdc.gov/vhf/ebola/history/chronology.html [accessed 15 October 2020].
- Feldmann H, Sanchez A, Geisbert TW. Filoviridae: marburg and ebola viruses. in Fields Virology: Sixth Edition. 2013; DOI:10.1007/978-1-4612-3900-0_30
- Sanchez A, Trappier SG, Mahy BWJ, et al. The virion glycoproteins of ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. U. S. A. (1996) doi:10.1073/pnas.93.8.3602.
- Mehedi M, Falzarano D, Seebach J, et al. A new ebola virus nonstructural glycoprotein expressed through RNA editing. Journal of Virology. 2011;85(11):5406–5414.
- Zhu W, Banadyga L, Emeterio K, et al. The roles of ebola virus soluble glycoprotein in replication, pathogenesis, and countermeasure development. Viruses. 2019;11(11):999.
- Muhlberger E, Weik M, Volchkov VE, et al. Comparison of the transcription and replication strategies of marburg virus and ebola virus by using artificial replication systems. Journal of Virology. 1999;73(3):2333–2342.
- Tchesnokov EP, Raeisimakiani P, Ngure M, et al. RNA-Dependent RNA Polymerase Complex of Ebola Virus. Sci Rep. 2018;8(1). DOI:10.1038/s41598-018-22328-3
- Bornholdt ZA, Noda T, Abelson D, et al. Structural rearrangement of ebola virus vp40 begets multiple functions in the virus life cycle. Cell. 2013;154(4):763–774..
- Harty RN, Brown ME, Wang G, et al. A PPxY motif within the VP40 protein of ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. (2000) doi:10.1073/pnas.250277297.
- Noda T, Sagara H, Suzuki E, et al. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76(10):4855–4865.
- Miller EH, Chandran K. Filovirus entry into cells – new insights. Curr Opin Virol. 2012;2(2):206–214.
- Jangra RK, Mittler E, Chandran K. Chapter 19: Filovirus entry into susceptible cells. in Biology and Pathogenesis of Rhabdo- and Filoviruses. 2014; DOI:10.1142/9789814635349_0019
- Huang Y, Xu L, Sun Y, et al. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10(2):307–316.
- Noda T, Halfmann P, Sagara H, et al. Regions in Ebola virus VP24 that are important for nucleocapsid formation. The Journal of Infectious Diseases. 2007;196(s2):S247-S250.
- Hoenen T, Groseth A, Kolesnikova L, et al. Infection of naïve target cells with virus-like particles: implications for the function of ebola virus VP24. J Virol. 2006;(14):7260–7264. DOI:10.1128/jvi.00051-06
- Mateo M, Carbonnelle C, Martinez MJ, et al. Knockdown of Ebola virus VP24 impairs viral nucleocapsid assembly and prevents virus replication. . The Journal of Infectious Diseases. 2011;204(suppl_3):S892-S896.
- Banadyga L, Hoenen T, Ambroggio X, et al. Ebola virus VP24 interacts with NP to facilitate nucleocapsid assembly and genome packaging. Sci Rep. 2017;7(1). DOI:10.1038/s41598-017-08167-8
- Watt A, Moukambi F, Banadyga L, et al. A novel life cycle modeling system for ebola virus shows a genome length-dependent role of VP24 in virus infectivity. Journal of Virology. 2014;88(18):10511–10524.
- Cong Q, Pei J, Grishin NV. Predictive and comparative analysis of Ebolaviru proteins. Cell Cycle. 2015;14(17). DOI:10.1080/15384101.2015.1068472
- Kuhn JH, Adachi T, Adhikari NKJ, et al. New filovirus disease classification and nomenclature. Nature Rev Microbiol. 2019;17(5):261–263.
- Team S, Branch SP, Division V, et al. Ebola haemorrhagic fever in sudan, 1976. Bull World Health Organ. 1978;56(2):247-270.
- Shoemaker T, MacNeil A, Balinandi S, et al. Reemerging sudan ebola virus disease in uganda, 2011. Emerg Infect Dis. 2012; DOI:10.3201/eid1809.111536..
- Towner JS, Sealy TK, Khristova ML, et al. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4(11):e1000212.
- Albariño CG, Shoemaker T, Khristova ML, et al. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in uganda and the democratic republic of the congo in 2012. Virology. 2013;442(2):97–100.
- Kratz T, Roddy P, Tshomba Oloma A, et al. Ebola virus disease outbreak in isiro, democratic republic of the congo, 2012: signs and symptoms, management and outcomes. PLoS One. 2015;10(6):e0129333.
- Burk R, Bollinger L, Johnson JC, et al. Neglected filoviruses. FEMS Microbiology Reviews. 2016;40(4):494–519.
- Le Guenno B, Formenty P, Wyers M, et al. Isolation and partial characterisation of a new strain of ebola virus. Lancet. 1995;345(8960):1271–1274.
- Formenty P, Hatz C, Le Guenno B, et al. Human infection due to ebola virus, subtype Côte d’Ivoire: clinical and biologic presentation. The Journal of Infectious Diseases. 1999;179(s1):S48-S53.
- Jahrling PB, Geisbert TW, Dalgard DW, et al. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet. 1990;335(8688):502–505.
- Barrette RW, Metwally SA, Rowland JM, et al. Discovery of swine as a host for the reston ebolavirus. Science. 2009;325(5937):204–206.
- Miranda MEG, Miranda NLJ. Reston ebolavirus in humans and animals in the philippines: a review. The Journal of Infectious Diseases. 2011;204(suppl_3):S757-S760.
- Cantoni D, Hamlet A, Michaelis M, et al. risks posed by reston, the forgotten ebolavirus. mSphere. 2016;1(6). DOI:10.1128/msphere.00322-16
- Centers for Disease Control. Update: Filovirus infection in animal handlers. Morb. Mortal. Wkly. Report. 39: 221. 1990;
- Goldstein T, Anthony SJ, Gbakima A, et al. The discovery of bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol. 2018; DOI:10.1038/s41564-018-0227-2.
- Forbes KM, Webala PW, Jääskeläinen AJ, et al. Bombali virus in mops condylu bat, kenya. Emerg Infect Dis. 2019;25(5). DOI:10.3201/eid2505.181666
- Haaskjold YL, Bolkan HA, Krogh KØ, et al. Clinical features of and risk factors for fatal Ebola virus disease, moyamba district, sierra leone, December 2014–February 2015. Emerging Infectious Diseases. 2016;22(9):1537–1544.
- Ji YJ, Duan XZ, Gao XD, et al. Clinical presentations and outcomes of patients with ebola virus disease in freetown, sierra leone. Infect Dis Poverty. 2016;5(1). DOI:10.1186/s40249-016-0195-9
- Vernet M-A, Reynard S, Fizet A, et al. Clinical, virological, and biological parameters associated with outcomes of ebola virus infection in macenta, guinea. JCI Insight. 2017;2(6). DOI:10.1172/jci.insight.88864
- Maganga GD, Kapetshi J, Berthet N, et al. Ebola virus disease in the democratic republic of congo. New England Journal of Medicine. 2014;371(22):2083–2091.
- Schieffelin JS, Shaffer JG, Goba A, et al. Clinical illness and outcomes in patients with ebola in sierra leone. N Engl J Med. 2014;371(22):2092–2100.
- Bah EI, Lamah M-C, Fletcher T, et al. Clinical presentation of patients with ebola virus disease in conakry, guinea. New England Journal of Medicine. 2015;372(1):40–47.
- Barry M, Traoré FA, Sako FB, et al. Ebola outbreak in conakry, guinea: epidemiological, clinical, and outcome features. Med Mal Infect. 2014;44(11–12):491–494.
- Barry M, Toure A, Traore FA, et al. Clinical predictors of mortality in patients with ebola virus disease. Clinical Infectious Diseases. 2015;60(12):1821–1824.
- Dallatomasina S, Crestani R, Sylvester Squire J, et al. Ebola outbreak in rural west africa: epidemiology, clinical features and outcomes. Trop Med Int Heal. 2015;20(4):448–454.
- Lado M, Walker NF, Baker P, et al. Clinical features of patients isolated for suspected ebola virus disease at connaught hospital, freetown, sierra leone: a retrospective cohort study. The Lancet Infectious Diseases. 2015;15(9):1024–1033.
- Yan T, Mu J, Qin E, et al. Clinical characteristics of 154 patients suspected of having ebola virus disease in the ebola holding center of jui government hospital in sierra leone during the 2014 ebola outbreak. Eur J Clin Microbiol Infect Dis. 2015;34(10):2089–2095.
- Bwaka MA, Bonnet MJ, Calain P, et al. Ebola hemorrhagic fever in kikwit, democratic republic of the congo: clinical observations in 103 patients. The Journal of Infectious Diseases. 1999;179(s1):S1-S7.
- McElroy AK, Erickson BR, Flietstra TD, et al. Ebola hemorrhagic fever: novel biomarker correlates of clinical outcome. Journal of Infectious Diseases. 2014;210(4):558–566.
- Velásquez GE, Aibana O, Ling, EJ, et al. Time from infection to disease and infectiousness for ebola virus disease, a systematic review. Clinl Infect Dis. 2015;61(7):1135–1140.
- Martines RB, Ng DL, Greer PW, et al. Tissue and cellular tropism, pathology and pathogenesis of ebola and marburg viruses. The Journal of Pathology. 2015;235(2):153–174.
- Reynard S, Journeaux A, Gloaguen E, et al. Immune parameters and outcomes during Ebola virus disease. JCI Insight. 2019;4(1). DOI:10.1172/jci.insight.125106
- Kerber R, Krumkamp R, Diallo B, et al. Analysis of diagnostic findings from the european mobile laboratory in guéckédou, guinea, March 2014 Through March 2015.Journal of Infectious Diseases. 2016;214(suppl 3):S250-S257.
- Kerber R, Krumkamp R, Korva M, et al. Kinetics of soluble mediators of the host response in ebola virus disease. The Journal of Infectious Diseases. 2018;218(suppl_5):S496-S503.
- Eisfeld AJ, Halfmann PJ, Wendler JP, et al. Multi-platform ’Omics analysis of human ebola virus disease pathogenesis. Cell Host Microbe. 2017;22(6):817–829.e8.
- Villinger F, Rollin P, Brar S, et al. Markedly Elevated Levels of Interferon (IFN)-σ, IFN-α, Interleukin (IL)-2, IL-10, and tumor necrosis factor-α associated with fatal ebola virus infection. The Journal of Infectious Diseases. 1999;179(s1):S188-S191.
- Gupta M, Mahanty S, Ahmed R, et al. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with Ebola virus secrete MIP-1α and TNF-α and inhibit poly-IC-induced IFN-α in vitro. Virology. 2001;284(1):20–25.
- Baize S, Leroy EM, Georges AJ, et al. Inflammatory responses in Ebola virus-infected patients. Clinical & Experimental Immunology. 2002;128(1):163–168.
- Wauquier N, Becquart P, Padilla C, et al. Human fatal zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis. 2010;4(10):e837.
- Bixler SL, Goff AJ. The role of cytokines and chemokines in filovirus infection. Viruses. 2015;7(10):5489–5507.
- Mandl JN, Feinberg MB Robust and sustained immune activation in human Ebola virus infection. Proc. Natl. Acad. Sci. U. S. A. (2015) doi:10.1073/pnas.1503864112.
- Ruibal P, Oestereich L, Lüdtke A, et al. Unique human immune signature of Ebola virus disease in Guinea. Nature. 2016;533(7601):100–104.
- Magrone T, Jirillo E. Sepsis: from historical aspects to novel vistas. Pathogenic and therapeutic considerations. Endocrine. Metab. Immune Disord. Drug Targets. 2019;19(4):490–502.
- McElroy AK, Akondy RS, Davis CW, et al. Human Ebola virus infection results in substantial immune activation. Proc. Natl. Acad. Sci. U. S. A. (2015) doi:10.1073/pnas.1502619112.
- Sakabe S, Sullivan BM, Hartnett JN, et al. Analysis of CD8+ T cell response during the 2013–2016 Ebola epidemic in west africa. Proc. Natl. Acad. Sci. U. S. A. (2018) doi:10.1073/pnas.1806200115.
- LaVergne SM, Sakabe S, Kanneh L, et al. Ebola-Specific CD8+ and CD4+ T-Cell responses in sierra leonean ebola virus survivors with or without post-ebola sequelae. . The Journal of Infectious Diseases. 2020;222(9):1488–1497.
- Baize S, Leroy EM, Georges-Courbot MC, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5(4):423–426.
- Geisbert TW, Hensley LE, Larsen T, et al. Pathogenesis of ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. . The American Journal of Pathology. 2003;163(6):2347–2370.
- Geisbert TW, Young HA, Jahrling PB, et al. Pathogenesis of ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am J Pathol. 2003;163(6):2371–2382.
- Bray M, Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. The International Journal of Biochemistry & Cell Biology. 2005;37(8):1560–1566.
- Ebihara H, Rockx B, Marzi A, et al. Host response dynamics following lethal infection of rhesus macaques with zaire ebolavirus. The Journal of Infectious Diseases. 2011;204(suppl_3):S991-S999.
- Warren T, Zumbrun E, Weidner JM, et al. Characterization of Ebola virus disease (EVD) in rhesus monkeys for development of EVD therapeutics. Viruses. 2020;12(1):92.
- Ryabchikova EI, Kolesnikova LV, Luchko SV. An analysis of features of pathogenesis in two animal models of Ebola virus infection. The Journal of Infectious Diseases. 1999;179(s1):S199-S202.
- Schnittler H-J, Feldmann H. Marburg and ebola hemorrhagic fevers: does the primary course of infection depend on the accessibility of organ-specific macrophages? Clinical Infectious Diseases. 1998;27(2):404–406.
- Cross RW, Mire CE, Borisevich V, et al. The domestic ferret (mustela putorius fur) as a lethal infection model for 3 species ebolavirus. J Infect Dis. 2016;214(4):565–569.
- Banadyga L, Dolan MA, Ebihara EH. Rodent-adapted filoviruses and the molecular basis of pathogenesis. J Mol Biol. 2016;428(17):3449–3466.
- Yamaoka S, Banadyga L, Bray M, et al. Small animal models for studying filovirus pathogenesis. in Marburg- and Ebolaviruses.Current Topics in Microbiology and Immunology. 2017; DOI:10.1007/82_2017_9
- Bray M, Davis K, Geisbert T, et al. A mouse model for evaluation of prophylaxis and therapy of ebola hemorrhagic fever. J Infect Dis. 1998;178(3):651–661.
- Chan M, Leung A, Griffin BD, et al. Generation and characterization of a mouse-adapted makona variant of ebola virus. Viruses. 2019;11(11):987.
- Connolly BM, Steele KE, Davis KJ, et al. Pathogenesis of experimental Ebola virus infection in guinea pigs. The Journal of Infectious Diseases. 1999;179(s1):S203-S217.
- Volchkov VE, Chepurnov AA, Volchkova VA, et al. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology. 2000;277(1):147–155.
- Subbotina E, Dadaeva A, Kachko A, et al. Genetic factors of ebola virus virulence in guinea pigs. Virus Res. 2010;153(1):121–133.
- Cross RW, Fenton KA, Geisbert JB, et al. Modeling the disease course of zaire ebolavirus infection in the outbred guinea pig. Journal of Infectious Diseases. 2015;212(suppl 2):S305-S315.
- Gibb TR, Bray M, Geisbert TW, et al. Pathogenesis of experimental ebola zaire virus infection in BALB/c mice. J Comput Pathol. 2001;125(4):233–242.
- Ebihara H, Zivcec M, Gardner D, et al. A syrian golden hamster model recapitulating ebola hemorrhagic fever. The Journal of Infectious Diseases. 2013;207(2):306–318.
- Rasmussen AL, Okumura A, Ferris MT, et al. Host genetic diversity enables ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;(6212):987–991. DOI:10.1126/science.1259595
- Bray M. The role of the Type I interferon response in the resistance of mice to filovirus infection. . Journal of General Virology. 2001;82(6):1365–1373.
- Brannan JM, Froude JW, Prugar LI, et al. Interferon α/β receptor-deficient mice as a model for ebola virus disease. J Infect Dis. 2015;212(suppl 2):S282-S294.
- Raymond J, Bradfute S, Bray M. Filovirus infection of STAT-1 knockout mice. The Journal of Infectious Diseases. 2011;204(suppl_3):S986-S990.
- Lüdtke A, Oestereich L, Ruibal P, et al. Ebola virus disease in mice with transplanted human hematopoietic stem cells. J Virol. 2015;89(8):4700–4704.
- Bird BH, Spengler JR, Chakrabarti AK, et al. Humanized mouse model of Ebola virus disease mimics the immune responses in human disease. J Infect Dis. 2015; DOI:10.1093/infdis/jiv538.
- Spengler JR, Prescott J, Feldmann H, et al. Human immune system mouse models of Ebola virus infection. Curr Opin Virol. 2017;25:90–96.
- Spengler JR, Saturday G, Lavender KJ, et al. Severity of disease in humanized mice infected with ebola virus or reston virus is associated with magnitude of early viral replication in liver. J Infect Dis. 2018;217(1):58–63.
- Lavender KJ, Williamson BN, Saturday G, et al. Pathogenicity of ebola and marburg viruses is associated with differential activation of the myeloid compartment in humanized triple knockout-bone marrow, liver, and thymus mice. The Journal of Infectious Diseases. 2018;218(suppl_5):S409-S417.
- Escudero-Pérez B, Rottstegge M, Lüdtke A, et al. Comparative pathogenesis of ebola virus and reston virus infection in humanized mice. JCI Insight. 2019;4(21). DOI:10.1172/jci.insight.126070.
- Wozniak DM, Lavender KJ, Prescott J, et al. The utility of human immune system mice for high-containment viral hemorrhagic fever research. Vaccines (Basel). 2020;8(1):98.
- Spengler JR, Lavender KJ, Martellaro C, et al. Ebola virus replication and disease without immunopathology in mice expressing transgenes to support human myeloid and lymphoid cell engraftment. J Infect Dis. 2016;214(suppl 3):S308-S318.
- Basler CF, Wang X, Mühlberger E, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U. S. A. (2000) doi:10.1073/pnas.220398297.
- Basler CF, Mikulasova A, Martinez-Sobrido L, et al. The ebola virus vp35 protein inhibits activation of interferon regulatory factor 3. Journal of Virology. 2003;77(14):7945–7956.
- Cardenas WB, Loo YM, Gale M Jr, et al. Ebola Virus VP35 Protein Binds Double-Stranded RNA and Inhibits Alpha/Beta Interferon Production Induced by RIG-I Signaling. J Virol. 2006;80(11):5168–5178..
- Feng Z, Cerveny M, Yan Z, et al. The VP35 protein of ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. . Journal of Virology. 2007;81(1):182–192.
- Schümann M, Gantke T, Ebola Virus ME. VP35 Antagonizes PKR Activity through Its C-Terminal Interferon Inhibitory Domain. J Virol. 2009;83(17):8993–8997.
- Leung DW, Prins KC, Borek DM, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. . Nature Structural & Molecular Biology. 2010;17(2):165–172.
- Leung DW, Prins KC, Basler CF, et al. Ebolavirus VP35 is a multifunctional virulence factor. Virulence. 2010;1(6):526–531.
- Luthra P, Ramanan P, Mire C, et al. Mutual antagonism between the ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe. 2013;14(1):74–84.
- Bale S, Julien JP, Bornholdt ZA, et al. Ebolavirus VP35 Coats the Backbone of Double-Stranded RNA for Interferon Antagonism. J Virol. 2013;87(18):10385–10388.
- Edwards MR, Liu G, Mire CE, et al. Differential regulation of interferon responses by ebola and marburg virus VP35 Proteins. Cell Rep. 2016;14(7):1632–1640.
- Hartman AL, Bird BH, Towner JS, et al. Inhibition of IRF-3 Activation by VP35 Is Critical for the High Level of Virulence of Ebola Virus. J Virol. 2008;82(6):2699–2704.
- Prins KC, Delpeut S, Leung DW, et al. Mutations Abrogating VP35 Interaction with Double-Stranded RNA Render Ebola Virus Avirulent in Guinea Pigs. . Journal of Virology. 2010;84(6):3004–3015.
- Woolsey C, Menicucci AR, Cross RW, et al. A VP35 mutant ebola virus lacks virulence but can elicit protective immunity to wild-type virus challenge. Cell Rep. 2019;28(12):3032–3046.e6.
- Reid SP, Leung LW, Hartman AL, et al. Ebola Virus VP24 Binds Karyopherin α1 and Blocks STAT1 Nuclear Accumulation. . Journal of Virology. 2006;80(11):5156–5167.
- Reid SP, Valmas C, Martinez O, et al. Ebola Virus VP24 Proteins Inhibit the Interaction of NPI-1 subfamily karyopherin α proteins with activated STAT1. Journal of Virology. 2007;81(24). DOI:10.1128/jvi.01097-07
- Mateo M, Reid SP, Leung LW, et al. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. Journal of Virology. 2010;84(2):1169–1175.
- Xu W, Edwards MR, Borek DM, et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014;16(2):187–200.
- Ebihara H, Takada A, Kobasa D, et al. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog. 2006;2(7):e73.
- Mateo M, Carbonnelle C, Reynard O, et al. VP24 Is a molecular determinant of Ebola virus virulence in guinea pigs. The Journal of Infectious Diseases. 2011;204(suppl_3):S1011-S1020.
- Cheresiz SV, Semenova EA, Chepurnov AA. Adapted Lethality: what We Can Learn from Guinea Pig-Adapted Ebola Virus Infection Model. Adv Virol. 2016;2016:1–14.
- Yen B, Mulder LCF, Martinez O, et al. Molecular Basis for Ebolavirus VP35 Suppression of Human Dendritic Cell Maturation. J Virol. 2014;88(21):12500–12510.
- Lubaki NM, Ilinykh P, Pietzsch C, et al. The lack of maturation of ebola virus-infected dendritic cells results from the cooperative effect of at least two viral domains. Journal of Virology. 2013;87(13):7471–7485.
- Lubaki NM, Younan P, Santos RI, et al. The ebola interferon inhibiting domains attenuate and dysregulate cell-mediated immune responses. PLoS Pathog. 2016;12(12):e1006031.
- Ilinykh PA, Lubaki NM, Widen SG, et al. Different Temporal Effects of Ebola Virus VP35 and VP24 Proteins on Global Gene Expression in Human Dendritic Cells. . Journal of Virology. 2015;89(15):7567–7583.
- Bosio CM, Aman MJ, Grogan C, et al. Ebola and marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188(11):1630–1638.
- Mahanty S, Hutchinson K, Agarwal S, et al. Cutting Edge: impairment of dendritic cells and adaptive immunity by ebola and lassa viruses. The Journal of Immunology. 2003;170(6):2797–2801.
- Okumura A, Pitha PM, Yoshimura A, et al. Interaction between ebola virus glycoprotein and host toll-like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J Virol. 2010;84(1):27–33.
- Okumura A, Rasmussen AL, Halfmann P, et al. Suppressor of Cytokine Signaling 3 Is an Inducible Host Factor That Regulates Virus Egress during Ebola Virus Infection. . Journal of Virology. 2015;89(20):10399–10406.
- Wahl-Jensen V, Kurz S, Feldmann F, et al. Ebola virion attachment and entry into human Macrophages profoundly effects early cellular gene expression. PLoS Negl Trop Dis. 2011;5(10):e1359.
- Olejnik J, Forero A, Deflubé LR, et al. Ebolaviruses associated with differential pathogenicity induce distinct host responses in human macrophages. . Journal of Virology. 2017;91(11). DOI:10.1128/jvi.00179-17
- Lai CY, Strange DP, Wong TAS, et al. Ebola virus glycoprotein induces an innate immune response in vivo via TLR4. Front Microbiol. 2017. DOI:10.3389/fmicb.2017.01571
- Dolnik O, Volchkova V, Garten W, et al. Ectodomain shedding of the glycoprotein GP of Ebola virus. Embo J. 2004;23(10):2175–2184.
- Escudero-Pérez B, Volchkova VA, Dolnik O, et al. Shed GP of Ebola Virus Triggers Immune Activation and Increased Vascular Permeability. PLoS Pathog. 2014;10(11):e1004509.
- Iampietro M, Santos RI, Lubaki NM, et al. Ebola virus shed glycoprotein triggers differentiation, infection, and death of monocytes through toll-like receptor 4 activation. The Journal of Infectious Diseases. 2018;218(suppl_5):S327-S334.
- Younan P, Iampietro M, Nishida A, et al. Ebola virus binding to Tim-1 on T lymphocytes induces a cytokine storm. MBio. 2017;8(5). DOI:10.1128/mBio.00845-17
- Geisbert TW, Pushko P, Anderson K, et al. Evaluation in nonhuman primates of vaccines against Ebola virus. Emerg Infect Dis. 2002;8(5):503–507.
- Pratt WD, Wang D, Nichols DK, et al. Protection of nonhuman primates against two species of ebola virus infection with a single complex adenovirus vector. . Clinical and Vaccine Immunology. 2010;17(4):572–581.
- Marzi A, Feldmann F, Hanley PW, et al. Delayed disease progression in cynomolgus macaques infected with ebola virus makona strain. Emerg Infect Dis. 2015;21(10):1777–1783.
- Warfield KL, Dye JM, Wells JB, et al. Homologous and heterologous protection of nonhuman primates by ebola and sudan virus-Like particles. PLoS One. 2015;10(3):e0118881.
- Marzi A, Yoshida R, Miyamoto H, et al. Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of ebola hemorrhagic fever. PLoS One. 2012;7(4):e36192.
- Hensley LE, Mulangu S, Asiedu C, et al. Demonstration of cross-protective vaccine immunity against an emerging pathogenic ebolavirus species. PLoS Pathog. 2010;6(5):e1000904.
- Falzarano D, Feldmann F, Grolla A, et al. Single immunization with a monovalent vesicular stomatitis virus-based vaccine protects nonhuman primates against heterologous challenge with bundibugyo ebolavirus. J Infect Dis. 2011;204(suppl_3):S1082-S1089.
- Mire CE, Geisbert JB, Marzi A, et al. Vesicular stomatitis virus-based vaccines protect nonhuman primates against bundibugyo ebolavirus. PLoS Neglected Tropical Diseases. 2013;7(12):e2600.
- Geisbert TW, Geisbert JB, Leung A, et al. Single-injection vaccine protects nonhuman primates against infection with marburg virus and three species of ebola virus. J Virol. 2009;83(14):7296–7304.
- Geisbert TW, Strong JE, Feldmann H. Considerations in the use of nonhuman primate models of ebola virus and marburg virus infection: table 1.. J Infect Dis. 2015;212(suppl 2):S91-S97.
- Sullivan NJ, Geisbert TW, Geisbert JB, et al. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424(6949):681–684.
- Reed DS, Lackemeyer MG, Garza NL, et al. Aerosol exposure to zaire ebolavirus in three nonhuman primate species: differences in disease course and clinical pathology. Microbes Infect. 2011;13(11):930–936.
- Alfson KJ, Avena LE, Beadles MW, et al. Particle-to-PFU ratio of ebola virus influences disease course and survival in cynomolgus macaques. J Virol. 2015;89(13):6773–6781.
- Alfson KJ, Avena L, Beadles M, et al. Intramuscular exposure of macaca fascicularis to low doses of low Passage- or cell culture-adapted sudan virus or ebola virus. Viruses. 2018;10(11):642.
- Kozak R, He A, Kroeker A, et al. Ferrets infected with bundibugyo virus or ebola virus recapitulate important aspects of human filovirus disease. J Virol. 2016;90(20):9209–9223.
- Kroeker A, He S, De La Vega M-A, et al. Characterization of Sudan Ebolavirus infection in ferrets. Oncotarget. 2017;8(28):46262–46272.
- Gupta M, MacNeil A, Reed ZD, et al. Serology and cytokine profiles in patients infected with the newly discovered Bundibugyo ebolavirus. Virology. 2012;423(2):119–124.
- Zaki SR, Shieh W-J, Greer P, et al. A novel immunohistochemical assay for the detection of Ebola virus in skin: implications for diagnosis, spread, and surveillance of Ebola hemorrhagic fever. The Journal of Infectious Diseases. 1999;179(s1):S36-S47.
- Zaki SR, Goldsmith CS. Pathologic features of filovirus infections in humans. Curr Top Microbiol Immunol. 1998. DOI:10.1007/978-3-642-59949-1_7
- Rollin PE, Bausch DG, Sanchez A. Blood chemistry measurements and d-dimer levels associated with fatal and nonfatal outcomes in humans infected with sudan ebola virus. The Journal of Infectious Diseases. 2007;196(s2):S364-S371.
- Hunt L, Gupta-Wright A, Simms V, et al. Clinical presentation, biochemical, and haematological parameters and their association with outcome in patients with Ebola virus disease: an observational cohort study. The Lancet Infectious Diseases. 2015;15(11):1292–1299.
- Gupta M, Goldsmith CS, Metcalfe MG, et al. Reduced virus replication, proinflammatory cytokine production, and delayed macrophage cell death in human PBMCs infected with the newly discovered Bundibugyo ebolavirus relative to Zaire ebolavirus. Virology. 2010;402(1):203–208.
- Boehmann Y, Enterlein S, Randolf A, et al. A reconstituted replication and transcription system for ebola virus reston and comparison with ebola virus zaire. Virology. 2005;332(1):406–417.
- Groseth A, Marzi A, Hoenen T, et al. The ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog. 2012;8(8):e1002847.
- Groseth A, Feldmann H, Theriault S, et al. RNA polymerase i-driven minigenome system for ebola viruses. Journal of Virology. 2005;79(7):4425–4433.
- Bosworth A, Dowall SD, Armstrong S, et al. Investigating the cellular transcriptomic response induced by the makona variant of ebola virus in differentiated THP-1 Cells. Viruses. 2019;11(11):1023.
- Kash JC, Muhlberger E, Carter V, et al. Global suppression of the host antiviral response by ebola- and marburgviruses: increased antagonism of the type i interferon response is associated with enhanced virulence. Journal of Virology. 2006;80(6):3009–3020.
- Hutchinson KL, Rollin PE. Cytokine and chemokine expression in humans infected with sudan ebola virus. The Journal of Infectious Diseases. 2007;196(s2):S357-S363.
- Xiang C, Young H, et al. Comparison of cellular gene expression in ebola-zaire and ebola-reston virus-infected primary human monocytes. . Nature Genetics. 1999;23(S3):82.
- Younan P, Ramanathan P, Graber J, et al. The toll-like receptor 4 antagonist eritoran protects mice from lethal filovirus challenge. MBio. 2017;8(2). DOI:10.1128/mBio.00226-17.
- Dilley KA, Voorhies AA, Luthra P, et al. The Ebola virus VP35 protein binds viral immunostimulatory and host RNAs identified through deep sequencing. PLoS One. 2017;12(6):e0178717.
- Guito JC, Albariño CG, Chakrabarti AK, et al. Novel activities by ebolavirus and marburgvirus interferon antagonists revealed using a standardized in vitro reporter system. Virology. 2017;501:147–165.
- Kimberlina CR Bornholdt ZA, Li S, et al. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. U. S. A. (2010) doi:10.1073/pnas.0910547107.
- Leung DW, Shabman RS, Farahbakhsh M, et al. Structural and functional characterization of reston ebola virus VP35 interferon inhibitory domain. Journal of Molecular Biology. 2010;399(3):347–357.
- Schwarz TM, Edwards MR, Diederichs A, et al. VP24-Karyopherin Alpha Binding Affinities Differ between Ebolavirus Species, Influencing Interferon Inhibition and VP24 Stability. J Virol. 2017;91(4). DOI:10.1128/jvi.01715-16
- Zhang APP, Bornholdt ZA, Liu T, et al. The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 2012;8(2):e1002550.
- Chakraborty S, Rao BJ, Asgeirsson B, et al. Correlating the ability of VP24 protein from ebola and marburg viruses to bind human karyopherin to their immune suppression mechanism and pathogenicity using computational methods. F1000Res. 2014;3:265.
- Pappalardo M, Juliá M, Howard MJ, et al. Conserved differences in protein sequence determine the human pathogenicity of ebolaviruses. Scientific Reports. 2016;6(1). DOI:10.1038/srep23743
- De Wit E, Munster VJ, Metwally SA, et al. Assessment of rodents as animal models for reston ebolavirus. J Infect Dis. 2011;204(suppl_3):968–972.
- Nehls J, Businger R, Hoffmann M, et al. Release of immunomodulatory ebola virus glycoprotein-containing microvesicles is suppressed by tetherin in a species-specific manner. Cell Rep. 2019;26(7):1841–1853.e6.
- Holmes EC, Dudas G, Rambaut A, et al. The evolution of Ebola virus: insights from the 2013-2016 epidemic. Nature. 2016;538(7624):193–200.
- Kugelman JR, Wiley MR, Mate S, et al. Monitoring of Ebola virus Makona evolution through establishment of advanced genomic capability in Liberia. Emerging Infectious Diseases. 2015;21(7):1135–1143.
- Hoenen T, Safronetz D, Groseth A, et al. Mutation rate and genotype variation of Ebola virus from Mali case sequences. Science. 2015;348(6230):117–119.
- Simon-Loriere E, Faye O, Faye O, et al. Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature. 2015;524(7563):102–104.
- Park DJ, Dudas G, Wohl S, et al. Ebola virus epidemiology, transmission, and evolution during seven months in sierra leone. Cell. 2015;161(7):1516–1526.
- Tong YG, Shi WF, Liu D, et al. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015; DOI:10.1038/nature14490.
- Diehl WE, Lin AE, Grubaugh ND, et al. Ebola virus glycoprotein with increased infectivity dominated the 2013–2016 Epidemic. Cell. 2016;167(4):1088–1098.e6.
- Urbanowicz RA, McClure CP, Sakuntabhai A, et al. Human adaptation of ebola virus during the west african outbreak. Cell. 2016;167(4):1079–1087.e5.
- Ladner JT, Wiley M, et al. Evolution and spread of ebola virus in liberia, 2014–2015. Cell Host Microbe. 2015;18(6):659–669.
- Dietzel E, Schudt G, Krähling V, et al. Functional characterization of adaptive mutations during the west african ebola virus outbreak. J Virol. 2017;91(2). DOI:10.1128/jvi.01913-16
- Olabode A, Gatherer D, Jiang X, et al. Identification of important amino acid replacements in the 2013-2016 Ebola virus outbreak. bioRxiv. 2016;2014:075168.
- Wang H, Shi Y, Song J, et al. Ebola viral glycoprotein bound to its endosomal receptor niemann-pick C1. Cell. 2016; DOI:10.1016/j.cell.2015.12.044.
- Ueda MT, Kurosaki Y, Izumi T, et al. Functional mutations in spike glycoprotein of Zaire ebolavirus associated with an increase in infection efficiency. Genes Cells. 2017;22(2):148–159.
- Wang MK, Lim SY, Lee SM, et al. Biochemical basis for increased activity of ebola glycoprotein in the 2013–16 Epidemic. Cell Host Microbe. 2017;21(3):367–375.
- Maximilian Fels J, Robert H. Bortz III, Tanwee Alkutkar, Eva Mittler, View ORCID ProfileRohit K. Jangra, Jennifer S. Spence, V. O. P. C. A glycoprotein mutation that emerged during the 2013–2016 Ebola virus epidemic alters proteolysis and accelerates membrane fusion. bioRxiv. 2020. DOI:10.1101/2020.07.13.201863
- Wong G, He S, Leung A, et al. Naturally Occurring Single Mutations in Ebola Virus Observably Impact Infectivity. . Journal of Virology. 2018;93(1). DOI:10.1128/jvi.01098-18
- Marzi A, Chadinah S, Haddock E, et al. Recently identified mutations in the ebola virus-makona genome do not alter pathogenicity in animal models. Cell Rep. 2018;23(6):1806–1816.
- Smither SJ, Eastaugh L, Ngugi S, et al. Ebola virus makona shows reduced lethality in an immune-deficient mouse model. J Infect Dis. 2016;214(suppl 3):S268-S274.
- Reisler RB, Yu C, Donofrio MJ, et al. Clinical laboratory values as early indicators of ebola virus infection in nonhuman primates. Emerging Infectious Diseases. 2017;23(8):1316–1324.
- Versteeg K, Menicucci AR, Woolsey C, et al. Infection with the makona variant results in a delayed and distinct host immune response compared to previous ebola virus variants. Sci Rep. 2017;7(1). DOI:10.1038/s41598-017-09963-y
- Jankeel A, Menicucci AR, Woolsey C, et al. Early transcriptional changes within liver, adrenal gland, and lymphoid tissues significantly contribute to ebola virus pathogenesis in cynomolgus macaques. . Journal of Virology. 2020;94(11). DOI:10.1128/jvi.00250-20
- Ivanov A, Ramanathan P, Parry C, et al. Global phosphoproteomic analysis of Ebola virions reveals a novel role for VP35 phosphorylation-dependent regulation of genome transcription. Cell Mol Life Sci. 2020;77(13):2579–2603.
- Cross RW, Speranza E, Borisevich V, et al. Comparative transcriptomics in ebola makona-infected ferrets, nonhuman primates, and humans. The Journal of Infectious Diseases. 2018; DOI:10.1093/infdis/jiy455.
- Wong G, Leung A, He S, et al. The makona variant of ebola virus is highly lethal to immunocompromised mice and immunocompetent ferrets. J Infect Dis. 2018; DOI:10.1093/infdis/jiy141.

