ABSTRACT
Dengue virus infection mainly causes dengue hemorrhagic fever (DHF) and/or dengue shock syndrome (DSS). However, ADE (antibody-dependent enhancement) is one of the main pathogenic factors, and its pathogenic mechanism has not been fully elucidated. Recently, with the development of high-throughput sequencing, an increased number of RNAs have been confirmed to play a vital regulatory role in the process of virus infection. However, there is a lack of research on dengue virus infection and ADE. In this study, we used RNA-Seq to detect differentially expressed RNAs (DE RNAs) profiles in mock-infected, DENV-3-infected, and ADE-infected THP-1 cells. Firstly, we found 69 circRNAs, 259 miRNAs, and 18 mRNAs were differentially expressed in THP-1 vs DENV-3. In THP-1 vs ADE, 94 circRNAs, 263 miRNAs, and 111 mRNAs were differentially expressed. In DENV-3 vs ADE, 68 circRNAs, 105 miRNAs, and 94 mRNAs were differentially expressed. Functional enrichment analysis of these DE RNAs mainly focused on immune system, viral infectious diseases, cytokine-cytokine receptor interactions, and NOD/RIG-I-like receptor signaling pathways. In DENV-3 vs ADE, notably, the expression of HBB was up-regulated, which was a Fcγ Receptor-mediated phagocytosis protein. Additionally, we predicted the encoding ability of DE circRNAs, and it was found that a small peptide was encoded by novel_circ_001562 and that its amino acid sequence was consistent with that of DDX60L, which is a class of interferon-stimulated genes. Finally, we constructed the ceRNA regulatory network pathway. Therefore, our study provides a new strategy for further investigation on DENV-host interactions.
Introduction
Dengue virus belongs to the Flavivirus genus of the Flaviviridae family. It is a type of arbovirus, which has been spread by Aedes albopictus and Aedes aegypti, and is widespread in tropical and subtropical regions [1]. Dengue virus is a type of enveloped, single positive-stranded RNA (+ssRNA) virus with a whole-genome length of approximately 10.7kb [Citation1]. The viral genome encodes a polyprotein with a whole length of 3,500 amino acids, which is cleaved by the host protease to form 10 proteins, including 7 nonstructural proteins and 3 structural proteins [Citation2]. The structural proteins mainly include Capsid protein (C protein), Envelope protein (E protein), and Pre-Membrane protein (PrM protein). The PrM protein can be cleaved by the host furin enzyme to form a mature M protein. The nonstructural proteins include nonstructural proteins 1 (NS1), nonstructural proteins 2A (NS2A), nonstructural proteins 2B (NS2B), nonstructural proteins 3 (NS3), nonstructural proteins 4 (NS4), nonstructural proteins 5A (NS5A), and nonstructural proteins 5B (NS5B). According to the antigenic difference of E protein, dengue virus can be divided into 4 sub-types: DENV-1, DENV-2, DENV-3, and DENV-4. Most of the infected people are asymptomatic or have only mild symptoms. About 25% of the infected people have fever (known as dengue fever) and abnormal blood biochemical indicators [Citation3]. Only a few infected people have severe symptoms, e.g., dengue hemorrhagic fever (DHF), dengue shock syndrome (DSS), vascular leakage syndrome, coagulation dysfunction, and multiple organ damage [Citation3]. Most severely infected people are more common in secondary infections or multiple infections. Because the first infection can stimulate the immune system to produce high levels of neutralizing antibodise, it can effectively protect against homotype and heterotype dengue virus infections over a period of time. However, as the neutralizing antibody titer drops to the sub-neutralizing antibody titer, it may promote homotype or heterotype dengue virus infection, thereby inducing immune system disorders, leading to serious illnesses [Citation4,Citation5]. This phenomenon is called ADE (antibody-dependent enhancement), and it was first proposed by Halstead and colleagues: when cross-antibody or sub-neutralizing antibody, the virus and antibody form a complex and promote the virus infection process through FCγR II. This phenomenon is found in dendritic cells, macrophages, and monocytes. Therefore, ADE is a receptor-mediated process that promotes viral infection and can escape the host’s antiviral immune response, leading to an imbalance of the inflammatory response, inflammatory cytokine storms, vascular leakage, and immune response imbalance that aggravate the process of disease [Citation6,Citation7]. With the development and application of RNA-Seq, it is found that about 80% of the genome of the host gene does not participate in the transcription and translation process, but it plays a vital regulatory mechanism in the interaction between virus and host [Citation8]. Recently, an increased number of non-coding RNAs have been reported on the regulation mechanism of various viral infections and interactions between host and virus [Citation9–12]. In flavivirus infections, some non-coding RNAs functions have also been reported. For example, microRNA-30e* inhibits the infection process of DENV-2 via regulating the NF-κB/IFN signaling pathway [Citation13]. lncRNA (long non-coding RNA) Malat1 is up-regulated during flavivirus infection, and regulates the host’s response to viral infection through the PERK signaling pathway [Citation14]. Single-cell sequencing of body fluid samples from patients infected with dengue virus revealed that some RNAs can predict the development of illness [Citation15]. In addition, microRNA-155 can regulate the process of antiviral and inflammatory response in West Nile virus infection [Citation16]. As a type of RNAs with special structure, circRNA mainly regulate the expression level of genes by adsorbing microRNAs to form a ceRNA regulatory network. This phenomenon is found in many diseases, such as tumor [Citation17,Citation18], cardiovascular disease [Citation19], infectious disease [Citation20], and obesity [Citation21]. However, at the level of RNAs, the pathogenesis of dengue virus infection and ADE, and the mechanism of the interaction between the virus and the host remain unclear. Therefore, we analyzed the DE RNA profiles and function in mock-infected, DENV-3-infected, and ADE-infected THP-1 cells and then analyzed whether circRNAs can encode small peptide amino acid sequences and their structures. The regulatory network of ceRNAs was built to supply a new strategy for investigation on the molecular mechanism of dengue virus infection and ADE.
Materials and methods
Cell and DENV-3
DENV-3 strain (GenBank number: MW426463) was from our department. RPMI-1640 medium containing streptomycin (100 μg/ml), penicillin (100 U/ml) and fetal bovine serum (10%) was used for THP-1 and C6/36 cell culture. THP-1 was used to establish DENV-3 infection and ADE models. C6/36 cells were used for virus amplification.
DENV-3 infection and ADE models in vitro
The Anti- Dengue- II PrM antibody (AB41473) and DENV-3 were used to establish DENV-3 infected THP-1 and ADE models at multiplicity of infection of 0.3, in accordance with our previous study [Citation22].
Western blotting (WB)
Mock-infected, ADE-infected, and DENV3-infected THP-1 cells were lysed by RIPA to extract total protein. The total protein was separated by SDS-PAGE, then transferred to PVDF membrane by electroporation, and blocked with skim milk (5%) at room temperature for 90 minutes. Then it was incubated with the primary antibody (Anti-DENV-III E protein antibody) overnight at four centigrade degree, and washed three times with PBST, 5 min each time. It was then incubated with the secondary antibody for 2 h under room temperature. Finally, the DENV-3 E protein signals can be visualized.
Immunofluorescence (IF)
The Anti-DENV-III E protein Antibody as primary antibody and Cy3-conjugated secondary antibody (AB97035) were used to stain the virus E protein in the cytoplasm, and DAPI (US EVERBRIGHT INC) was used to stain the nucleus. Finally, the DENV-3 E protein signal and the nucleus were observed under a confocal microscope.
RNA extraction and RNA analysis
RNA extraction
Intracellular total RNA was extracted using Trizol (Invitrogen) 48 h after mock-infection, DENV3-infection, and ADE-infection, in accordance with the manufacturer’s instruction.
Detection of viral nucleic acid copy number by qRT-PCR
According to our previous experiment [Citation22], DENV-3-qRT-PCR Primers and Plasmid-DENV-3-qRT-PCR Primers (Table S1.) were designed to target the DENV-3 PrM and DENV-3 PrM within the standard, respectively. Then, the number of viral nucleic acid copies was converted according to the standard curve. The qRT-PCR reagents are TB Green Premix Ex TaqTM II (TAKARA RR820A), in accordance with the manufacturer’s instruction.
Identification of circRNAs and DE circRNAs
The intracellular total RAN was digested to degrade linear RNA with RNaseR, and then purified using RNeasy MinElute Cleanup Kit (Qiagen) . These digestive products were sequenced by PCR amplification, and then RNAs sequence was compared with Find_Circ. Differential analysis was performed for all CircRNAs using edgeR package with a fold change ≥2. p value <0.05 indicated DE CircRNAs.
ORFs Prediction and IRES prediction
The two software, which are cORF pipeline [Citation23] script and IRES finder [Citation24], were used to predict ORF and IRES to determine if these DE CircRNAs can encode the polypeptide.
Identification of miRNA and differentially expressed miRNAs
All of the clean data were compared with miRBase database (Release 22). All miRNAs were analyzed using edgeR package with a fold change ≥2, and p value <0.05 indicated DE miRNAs.
Functional enrichment analysis
Gene Ontology Functional Enrichment Analysis (GO, http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes Functional Enrichment Analysis (KEGG, https://www.kegg.jp/)
Construction of differentially expressed “circRNAs-miRNAs-mRNAs” regulatory network
In order to foretell DE miRNAs sponge with DE circRNAs and mRNAs, the miRTarBase (version 6.1) was used to find differentially expressed miRNAs interacting with DE mRNAs and circRNAs. The correlation of DE circRNAs-miRNAs and DE miRNAs-mRNA can be visualized via Cytoscape (https://cytoscape.org/).
Differentially expressed circRNAs, microRNAs and mRNAs were identified by qRT-PCR analysis
In order to verify the accuracy of RNA-Seq. Fisrtly, we using PrimeScriptTM reagent kit with gDNA Eraser (TAKARA RR047A) and PCR instrument (Bio-Rad) to reverse transcription reaction, according to the instructions. Then, we using TB Green Premix Ex TaqTM II (TAKARA RR820A) and Fluorescence quantitative PCR instrument (Bio-Rad) to detect the RNA expression level, according to the instructions.The RT-qPCR was used to investigate the relative levels of DE RNAs. The primers for those DE RNAs and GAPDH are shown in Table S1. MicroRNAs needs to replace the random primers in Takara RR047A with specific primers (Table S1) for specific stem ring detection to reverse transcription reaction.
Statistical analysis
SPSS17.0 software was used to analyze the mean value difference after three independent repeated trials in different groups. Through one-way ANOVA of multiple groups, it was determined that the difference was statistically significant when p < 0.05. Single, double and three asterisks, and * indicate statistical significance (* p < 0.05; * p < 0.01; * p < 0.001).
Results
Establishment of a model of ADE in DENV-3 infected THP-1 cells
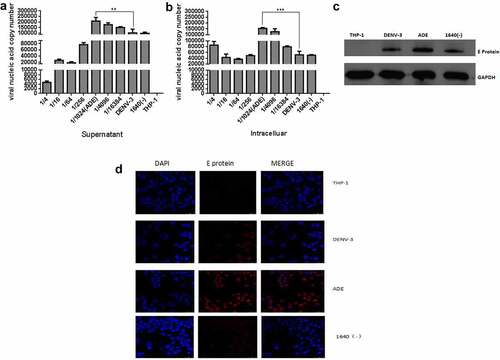
Currently, it is considered that ADE is mediated by FCγR [Citation25], and ADE models of dengue virus infection have been successfully established in THP-1, U937, and K562 cells [Citation26]. Based on our previous research [Citation22], Anti-DENV-II PrM antibody and DENV-3 were used to set up DENV-3 infection and ADE models in THP-1 cells. At 48 h post-infection, the qRT-PCR was used to detect the DENV-3 genome RNA in the supernatant and cells, and the Western blotting and Immunofluorescence were used to detect the intracellular DENV-3 E protein. As shown in , different dilutions of anti-DENV-II PrM antibodies promoted or inhibited DENV-3 infection compared with DENV-3 direct infection in THP-1 cells. When the anti-DENV-II PrM was diluted by 1/1,024, the copy number of viral nucleic acid in the cell and supernatant was significantly higher compared to DENV-3 direct infection and other dilutions. The same results can also be observed in . At a dilution of 1/1,024, the DENV-3 E protein was found to be significantly higher than that of direct infection with DENV-3. Therefore, DENV-3 can infect THP-1, and the anti-DENV-II PrM antibody at a dilution of 1/1,024 can establish the ADE model in THP-1.
Figure 1. Establishing a model of ADE in DENV-3 infected THP-1 cells
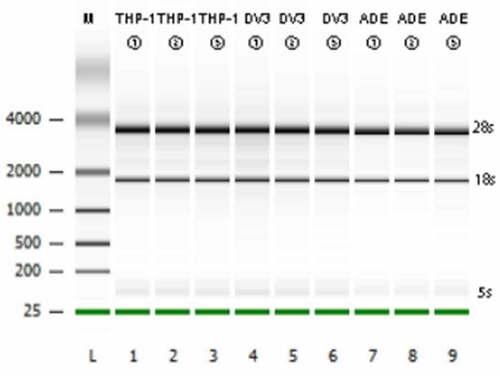
Transcriptome sequencing data quality assessment
Before proceeding with RNA-Seq, agarose gel electrophoresis was used to detect the integrity of the total RNA extracted from the cells. According to Bioanalyzer plot, as shown in ), S28, S18, and S5 bands were clearly visible, indicating that the integrity of the RNA was good and fulfilled the requirements of sequencing. The transcriptome sequencing on the IIIumina 6000 harvested at least 6.0Gb of clean data per sample, which can be used for further analysis after quality control. The clean data of all samples were above 99.77% (), and the GC content of the cleaning data of all samples were 46.6–47.58% (). These high-quality clean data were compared with the reference human genome (RHG, Ensembl_release102. GenBank number: GCA-000001405.28). Approximately 96.22% to 97.05% of the clean reads (CRs) were successfully matched to the RHG, and 92.69% to 93.8% of the CRs were uniquely matched to the RHG (). Thus, it can be seen that the completeness and reliability of the sequencing results are high, and these results can be used for further research.
Table 1. Transcriptome sequencing data quality assessment
Analysis of DE circRNAs in mock-infected, DENV3-infected, and ADE-infected THP-1 cells
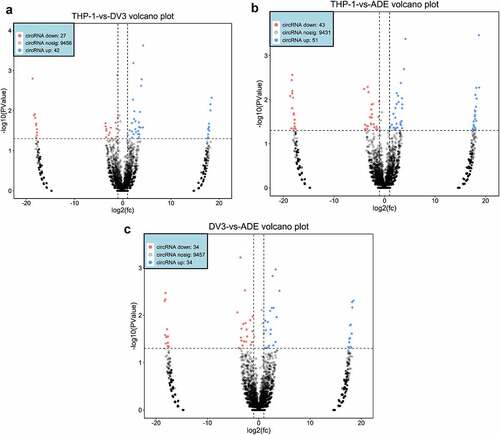
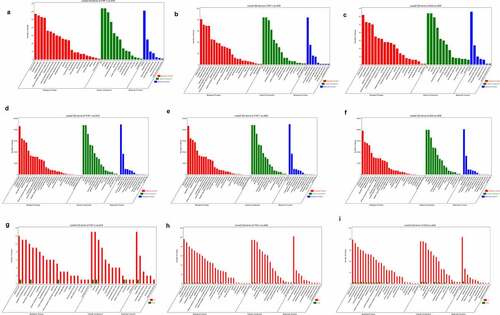
Using rRNA deletion samples from the RNA of mock-infected, DENV3-infected, and ADE-infected THP-1 cells, circRNA sequencing was performed. An average of about 79 million, 80 million, and 89 million CRs were found in the THP-1, DENV-3, and ADE groups, respectively (). 78.9% of DE circRNAs were exons, 4.04% were introns, and 1.09% were located at the intergenic region (Figure S1A, Supplementary material 1). Then, it was found that the size of most DE circRNAs was 100–3,000 nt. The predicted splicing lengths of DE circular RNAs were 100–1,000nt (Figure S1B, Supplementary material 1). These differentially expressed circRNAs were located at chromosomes, such as chromosome 17, chromosome 5, chromosome 3, chromosome 2, and chromosome 1 (Figure S1C, Supplementary material 1). A total of 69, 94, and 68 DE circRNAs were identified in THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE, respectively. Among those, in THP-1 vs DENV-3 ()), 42 DE circRNAs expressions were increased and 27 DE circRNAs expressions were reduced. In THP-1 vs ADE ()), 51 DE circRNAs expressions were increased and 43 DE circRNAs expressions were reduced. In DENV-3 vs ADE ()), 34 DE circRNAs expressions were increased and 34 DE circRNAs expressions were decreased. In addition, we also performed a heatmap analysis on all DE circRNAs (Figure S1D, Supplementary material 1). In DENV-3 vs ADE, one half of those dysregulated DE circRNAs were down-regulated and the other half were up-regulated.
Analysis of DE mRNAs and DE miRNAs in mock-infected, DENV-3-infected, and ADE-infected THP-1 cells
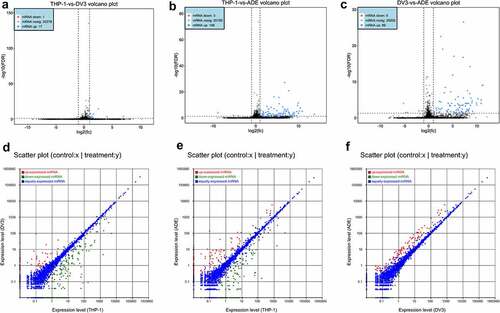
We detected 18, 111 and 94 DE mRNAs in THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE, respectively. Among those, 17 DE mRNAs were up-regulated in THP-1 vs DENV-3, and 1 was down-regulated ()). A total of 108 DE mRNAs were up-regulated in THP-1 vs ADE, and 3 were down-regulated ()). In DENV-3 vs ADE, 89 DE mRNAs expressions were increased and 5 were reduced ()). In addition, a heatmap analysis was performed on all DE mRNAs, and the results are shown in Figure S2A (Supplementary material 1) . Therefore, in DENV-3 infection and ADE, most of the DE mRNAs were up-regulated. Additionally, the sequencing data showed 252, 254, and 96 DE miRNAs were abnormally expressed in THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE, respectively. Among THP-1 vs DENV-3, 68 DE miRNAs expressions were increased and 184 DE miRNAs expressions were reduced ()). In THP-1 vs ADE, 123 DE miRNAs expressions were raised, and 131 DE miRNAs expressions were lowered ()). In DENV-3 vs ADE, 92 DE miRNAs expressions were enhanced and 4 DE miRNAs expressions were decreased ()). In addition, a heatmap analysis was also performed on all DE miRNAs (Figure S2B, Supplementary material 1). Compared with the DENV-3 vs ADE, most of dysregulated DE miRNAs were down-regulated in the THP-1 vs DENV-3. In the THP-1 vs ADE group, the number of DE miRNAs up-regulated and down-regulated was basically the same.
Figure 4. Differentially expressed mRNAs and miRNAs profiles
Functional enrichment analysis of the DE RNAs
In order to explore the biological function of DE RNAs in mock-infected, DENV-3-infected, and ADE-infected THP-1 cells, we tested the parental genes of differentially expressed RNAs through functional analysis. According to the results of GO analysis, DE circRNAs were mainly enriched in various biological processes (), such as “metabolic process”, “biological regulation” and “response to stimulation”. Additionally, the enrichment analysis of KEGG pathway showed that many DE circRNAs were mainly enriched in the “immune system” and “immune diseases”, “infectious diseases: viruses” and “cell growth and death” (Figure S3A-S3B, Supplementary material 1). Genes related to the “immune system” were up-regulated in DENV-3 vs ADE. These data indicated that differentially expressed circRNAs were mainly related to “infectious diseases: viral” and “host immune response”. According to the results of GO analysis, differentially expressed miRNAs were mainly enriched in a variety of biological processes, including “cellular processes”, “metabolic processes”, “biological regulation”, “response to stimuli” and “immune system” (. In addition, KEGG pathway enrichment analysis showed that DE miRNAs were mainly enriched in “apoptosis”, “cytokine-cytokine-receptor interaction” and “NOD/RIG-I-like receptor signaling pathway” (Figure S3D-S3F, Supplementary material 1). These DE miRNAs were mainly involved in “apoptosis” and “NOD/RIG-I-like receptor signaling pathways” in DENV-3 vs ADE. The parental genes of differentially expressed mRNAs were mainly enriched in different biological processes, such as “biological regulation”, “cellular processes”, and “immune system processes” (. Moreover, many differentially expressed mRNAs were mainly enriched in “Fcγ receptor-mediated phagocytosis (such as HBB)”, “complement and coagulation cascade”, “ECM receptor interaction” and “cell growth and death” according to the results of KEGG pathway enrichment analysis (Figure S5G-S5I, Supplementary material 1). These data confirm that the interaction between dengue virus and host is a complex mechanism, and those RNAs participate in the antiviral mechanism through various ways.
Figure 5. GO and KEGG analysis of DE RNAs
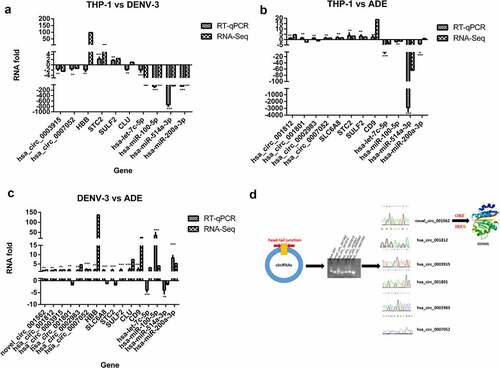
Quantitative real-time PCR analysis and prediction of the small peptide-encoding ability of the DE circRNAs of host genes
In order to confirm the results of RNA-Seq, we used qRT-PCR to verify the results of selected 10, 12, and 16 DE RNAs in THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE, respectively. As shown in (, RNA-Seq results were verified by RT-qPCR. The hand-tail joint sequence of DE circRNAs was confirmed by PCR and Sanger sequencing ()). Then, we predicted the ORF (open reading frame) and the IRES (internal ribosome entry site) to confirm whether these 6 differentially expressed circRNAs (novel_circ_001562, hsa_circ_001812, hsa_circ_0003915, hsa_circ_001801, hsa_circ_0002983, and hsa_circ_0007052) can encode small peptides. Except hsa_circ_0002983 (Supplementary material 2), other DE circRNAs can encode small peptides. Among these, the new_circ_001562 was predicted to encode a small peptide. BLAST was performed on the amino acid sequence of the small peptide in NCBI, and then its spatial structure was predicted ()). It was found that its amino acid sequence was consistent with DDX60L, which belonged to interferon-stimulating genes [Citation27]. In summary, these circRNAs may be involved in the interaction between DENV-3 and the host by regulating the host immune system and encoding polypeptide.
Figure 6. Quantitative real-time PCR analysis and Prediction of small peptide coding ability of circRNA encoded by host gene
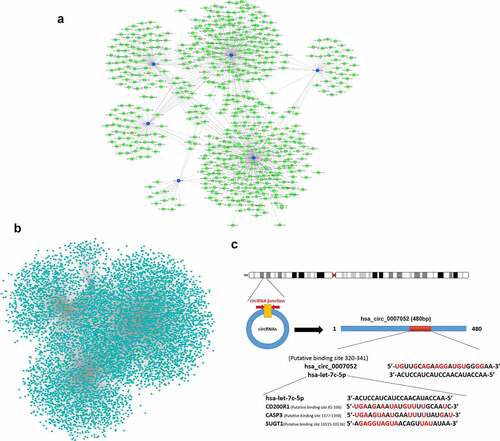
The regulation network of differentially expressed miRNAs, circRNAs, and mRNAs
It has been revealed that miRNAs can be competitively sponged by circRNAs, and gene expression is indirectly regulated. To examine the involvement of 6 DE circRNAs (novel_circ_001562, hsa_circ_001812, hsa_circ_0003915, hsa_circ_001801, hsa_circ_0002983, and hsa_circ_0007052), an integrative analysis was performed on the interaction of circRNAs and the target miRNAs. The analysis of miRNAs binding sites in 6 DE circRNAs revealed 661 miRNAs binding sites (), Supplementary material 3), including the site in hsa_circ_0007052 that can bind with hsa-let-7 c-5p. Next, 19,381 target mRNAs of the 4 differentially expressed miRNAs were predicted, and a miRNAs-mRNAs interaction network was constructed (), Supplementary material 4), and it was found that hsa-let-7 c-5p can bind with CD200R1, CASP3, SUGT1, etc. CircRNAs (hsa_circ_0007052) may act as ceRNAs, and miRNAs (hsa-let-7 c-5p) is sequestered to interrupt their binding and targeting of mRNAs (CD200R1, CASP3, SUGT1, etc.) ()). Therefore, circRNA-miRNA-mRNA interactions could play significant roles in the DENV-3-host interaction.
Figure 7. Differentially expressed CircRNAs- miRNAs- mRNAs interaction network
Discussion
The function of non-coding RNA has been mainly elucidated in cancer and disease progression [Citation17,Citation18,Citation21]. However, the expression profile, function and mechanism of these non-coding RNAs in viral infections are still unclear. Especially in the process of DENV-3 infection and ADE infection, the characteristics and potential role of non-coding RNA have not been explored. In the current study, the profiles of differentially expressed mRNAs, miRNAs and circRNAs in mock-infected, DENV-3-infected, and ADE-infected THP-1 cells were analyzed. It was found that DENV-3 infection significantly affected the RNA expression profile. In THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE, a total of 69, 94 and 68 DE circRNAs were identified respectively. Among these, in THP-1 vs DENV-3, 42 DE circRNAs expressions were increased and 27 were decreased. In THP-1 vs ADE, 51 DE circRNAs expressions were increased and 43 were decreased. In DENV-3 vs ADE, 34 DE circRNAs expressions were increased and 34 were reduced. Then, we detected 18, 111 and 94 DE mRNAs in THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE, respectively. Among these, most DE mRNAs (94.4%) were up-regulated, and only one was down-regulated in THP-1 vs DENV-3. Similarly, most of DE mRNAs were up-regulated in THP-1 vs ADE (97.3%) and DENV-3 vs ADE (94.7%). There were 252, 254 and 96 DE miRNAs expression abnormalities in THP-1 vs DENV-3, THP-1 vs ADE, and DENV-3 vs ADE respectively. Among THP-1 vs DENV-3, 68 DE miRNAs expressions were increased and 184 DE miRNAs expressions were decreased. In THP-1 vs ADE, the expression of 123 DE miRNAs was up-regulated, and the expression of 131 DE miRNAs was down-regulated. In DENV-3 vs ADE, 92 DE miRNAs were up-regulated and the expressions of 4 DE miRNAs were decreased. It was implied that the regulating mechanism of DE RNAs in the interaction between DENV-3 and host cells may be complex. The activation of innate immunity plays a vital role in the antiviral mechanism, and it is closely related to the regulation of a variety of molecules including RNA. It has been recently reported that some RNAs inhibit host antiviral immunity [Citation11,Citation28,Citation29]. Consistently, the DE circRNAs via GO and KEGG pathways show that these genes are mainly involved in “immune system processes” and “infectious diseases: viruses”. DE mRNAs were found to be involved in “Fcγ receptor-mediated phagocytosis” (such as HBB) and “complement and coagulation cascade”, DE miRNAs were found to be mainly involved in “cytokine-cytokine receptor interaction” and “NOD/RIG-I-like receptor signaling pathway”. In the DENV-3 vs ADE, the expression of HBB was upregulated. Through functional enrichment analysis, it was confirmed that HBB is an Fcγ receptor-mediated phagocytosis protein. This result is consistent with the Fcγ receptor hypothesis of ADE. These data indicate that DE RNAs may regulate DENV-3 infection and ADE infection via antiviral responses, host immune signaling pathways and Fcγ receptor-mediated phagocytosis to promote infection of the dengue virus. Many studies have shown that circRNAs can act as miRNAs sponges to indirectly regulate gene expression. Therefore, we constructed a ceRNA network to reveal the specific role and mechanism of DE circRNAs in DENV-3 and ADE infection. The integration analysis of ceRNA revealed the regulatory function of circRNAs and its specific interaction with mRNAs through the regulation axis of circRNA-miRNA-mRNA in the natural immune response of host cells to DENV-3. We found that circRNA-miRNA-mRNA axis may be involved in the regulation of DENV-3 and ADE infection, such as the hsa_circ_0007052 – hsa-let-7 c-5p – CD200R1/CASP3/ SUGT1 pathway. The CD200R1 signaling pathway can regulate host-pathogen interactions [Citation30], e.g., promoting HSV-1 infection and inflammation through Toll-like receptor 2 (TLR2) [Citation31]. CASP3 inhibits type I interferon through the cleavage of IRF3 and MAVS [Citation32]. SUGT1 regulates HIV-1 infection by stabilizing the positive end of microtubules [Citation33]. Thus, it is speculated that the disorder of differentially expressed circRNAs may be associated with the replication of DENV-3 via antagonizing the inhibition of mRNAs by miRNAs. In addition, many studies have confirmed that circRNAs encode small peptides [Citation34–38]. Considering this evidence, we performed ORF and IRS predictions for 6 differentially expressed circRNAs (novel_circ_001562, hsa_circ_001812, hsa_circ_0003915, hsa_circ_001801, hsa_circ_0002983, hsa_circ_0007052), and found 5 differentially expressed circRNAs (novel_circ_001562, hsa_circ_001812, hsa_circ_0003915, hsa_circ_001801, and hsa_circ_0007052) to determine their encoding potential. Then, we performed BLAST alignment and spatial structure prediction on the amino acid sequence of the small peptide encoded by the novel_circ_001562, and found that it was highly similar to DDX60L, which is an interferon-stimulated gene that can inhibit the replication of hepatitis C virus [Citation27]. Thus, our report firstly reveals DE RNAs profiling and a network of ceRNAs in DENV-3 infection and ADE infection that are involved in the regulation of DENV-3-host interactions, and innovatively investigates mechanisms underlying DENV-3 infection and ADE infection.
Authors’ contributions
Mingwang Long and Qiangming Sun designed the research.Yue Pan, Junying Chen, Kai feng, Han Wang, and Lingmei Yan performed a literature review.Fan Jia, Daiying Li, Xiaodan Wang, Xuelei Ning, Lijuan Qiu, and Juan Zhang provided the study materials. Mingwang Long performed the experiment, wrote the manuscript and analyzed the data.Qiangming Sun performed the article revision.
Supplemental Material
Download Zip (6.7 MB)Acknowledgments
We thank AJE for language revision of this manuscript and Gene Denovo Biotechnology Co.(Guangzhou, China) for their technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
In this study, those data are available from the corresponding author based on reasonable requests.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Slon Campos JL, Mongkolsapaya J, Screaton GR. The immune response against flaviviruses. Nat Immunol. 2018 Nov;19(11):1189–1198. Epub 2018 Oct 17. PMID: 30333606.
- Lindenbach BD, Heinz-Jurgen T, Rice CM. Flaviviridae: the viruses and their replication. In: editors, Fields BN, Knipe DM, Howley PM. Fields’ Virology. 5th edn ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007.
- WHO. Special Programme for Research and Training in Tropical Diseases. Dengue: guidelines for diagnosis, treatment, prevention and control: new edition. Geneva: World Health Organization; 2009.
- Halstead SB, Nimmannitya S, Cohen SN. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J Biol Med. 1970;42:311–328.
- Halstead SB, O’Rourke EJ. Dengue viruses and mononuclear phagocytes. Infection enhancement by non-neutralizing antibody. J Exp Med. 1977;146(1):201–217.
- Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11(8):532–543.
- Yacoub S, Wertheim H, Simmons CP, et al. Microvascular and endothelial function for risk prediction in dengue: an observational study. Lancet. 2015;385(suppl 1):S102.
- Lu S, Zhu N, Guo W, et al. Hou W. RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection. Front Cell Infect Microbiol. 2020 Mar 13;10:97. PMID: 32232013; PMCID: PMC7083127.
- Wang P, Xu J, Wang Y, et al. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017 Nov 24;358(6366):1051–1055. Epub 2017 Oct 26. PMID: 29074580.
- Cx L, Li X, Nan F, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019 May 2;177(4):865–880.e21. Epub 2019 Apr 25. PMID: 31031002.
- Pedersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922.
- Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381.
- Zhu X, He Z, Hu Y, et al. MicroRNA-30e* suppresses dengue virus replication by promoting NF-κB-dependent IFN production. PLoS Negl Trop Dis. 2014;8(8):e3088.
- Sankar B, Sudhanshu V. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. [J] .Sci Rep. 2015;5:17794.
- Zanini F, Robinson Makeda L, Croote D, et al. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc Natl Acad Sci U S A. 2018;115(52):E12363–E12369.
- Natekar JP, Rothan Hussin A, Arora K, et al. Cellular microRNA-155 Regulates Virus-Induced Inflammatory Response and Protects against Lethal West Nile Virus Infection. Viruses. 2019;12(1):9. undefined
- Ma M, Xu H, Liu G, et al. Metabolism-induced tumor activator 1 (MITA1), an energy stress-inducible long noncoding RNA, promotes hepatocellular carcinoma metastasis. Hepatology. 2019;68:215-230.
- Jiang S, Zhang LF, Zhang HW, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31(8):1985–1998.
- Gao F, Kataoka M, Liu N, et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat Commun. 2019;10(1):1802.
- Xia P, Wang S, Ye B, et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cgas-mediated exhaustion. Immunity. 2018;48(4):688–701.e7.
- Liu C, Ge HM, Liu BH, et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc Natl Acad Sci USA. 2019;116(15):7455–7464.
- Huang X, Yue Y, Li D, et al. Antibody-dependent enhancement of dengue virus infection inhibits RLR-mediated Type-I IFN-independent signalling through upregulation of cellular autophagy. Sci Rep. 2016 Feb 29;6(1):22303. PMID: 26923481; PMCID: PMC4770412.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs[J]. Mol Cell. 2017;66(1):9–21. e7.
- Zhao J, Wu J, Xu T, et al. IRESfinder: identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features[J]. Journal of genetics and genomics= Yi chuan xue bao. 2018;45(7):403.
- Moi ML, Takasaki T, Saijo M, et al. Determination of antibody concentration as the main parameter in a dengue virus antibody-dependent enhancement assay using FcgammaR-expressing BHK cells. Arch Virol. 2014;159(1):103–116.
- Jin X, Block OT, Rose R, et al. Dengue vaccine development and dengue viral neutralization and enhancement assays. Antivir Ther. 2009;14(6):739–749.
- Grünvogel O, Esser-Nobis K, Reustle A, et al. DDX60L Is an Interferon-Stimulated Gene Product Restricting Hepatitis C Virus Replication in Cell Culture. J Virol. 2015 Oct;89(20):10548–10568. Epub 2015 Aug 12. PMID: 26269178; PMCID: PMC4580188.
- Edersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;341:789–792. , 449: 919–922.2013
- Chen YG, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intronidentity. Mol Cell. 2017;67(2):228–238.e5.
- Ca V, Soberman RJ. The CD200-CD200R1 inhibitory signaling pathway: immune regulation and host-pathogen interactions. Adv Immunol. 2014;121:191–211. PMID: 24388216; PMCID: PMC4617684
- Rj S, MacKay CR, Vaine CA, et al.CD200R1 supports HSV-1 viral replication and licenses pro-inflammatory signaling functions of TLR2.PLoS One.2012;7(10):e47740. Epub 2012 Oct 17 PMID: 23082204; PMCID: PMC3474780.
- Ning X, Wang Y, Jing M, et al. Apoptotic Caspases Suppress Type I Interferon Production via the Cleavage of cGAS, MAVS, and IRF3. Mol Cell. 2019 Apr 4;74(1):19–31.e7. Epub 2019 Mar 13. PMID: 30878284.
- Allouch A, Di Primio C, Paoletti A, et al. SUGT1 controls susceptibility to HIV-1 infection by stabilizing microtubule plus-ends. Cell Death Differ. 2020 Dec;2712:3243–3257.
- Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018 Mar;37(13):1805–1814. Epub 2018 Jan 18. PMID: 29343848.
- Chen H, Liu Y, Li P, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2019Apr1;111(4):435. PMID: 29986033.
- Xia X, Li X, Li F, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019 Aug 30;18(1):131. Erratum in: Mol Cancer. 2019 Oct 29;18(1):149.PMID: 31470874; PMCID: PMC6716823.
- Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018 Oct 26;9(1):4475. 30367041; PMCID: PMC6203777.
- Li J, Ma M, Yang X, et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol Cancer. 2020 Sep 11;19(1):142. PMID: 32917240; PMCID: PMC7488427.