ABSTRACT
In many Gram-positive bacteria, the transpeptidase enzyme sortase A (SrtA) anchors surface proteins to cell wall and plays a critical role in the bacterial pathogenesis. Here, we show that in Staphylococcus aureus, an important human pathogen, the SrtA is phosphorylated by serine/threonine protein kinase Stk1. S. aureus SrtA can also be phosphorylated by small-molecule phosphodonor acetyl phosphate (AcP) in vitro. We determined that various amino acid residues of S. aureus SrtA are subject to phosphorylation, primarily on its catalytic site residue cysteine-184 in the context of a bacterial cell lysate. Both Stk1 and AcP-mediated phosphorylation inhibited the enzyme activity of SrtA in vitro. Consequently, deletion of gene (i.e. stp1) encoding serine/threonine phosphatase Stp1, the corresponding phosphatase of Stk1, caused an increase in the phosphorylation level of SrtA. The stp1 deletion mutant mimicked the phenotypic traits of srtA deletion mutant (i.e. attenuated growth where either haemoglobin or haem as a sole iron source and reduced liver infections in a mouse model of systemic infection). Importantly, the phenotypic defects of the stp1 deletion mutant can be alleviated by overexpressing srtA. Taken together, our finding suggests that phosphorylation plays an important role in modulating the activity of SrtA in S. aureus.
Introduction
Staphylococcus aureus is an important pathogen that causes life-threatening infections and poses a major public health threat due to the emergence of multidrug resistance [Citation1]. Potential alternative approaches to the treatment of S. aureus infection, particularly those caused by Methicillin-resistant S. aureus (MRSA), are agents that target S. aureus virulence [Citation2–5] and may include the inhibition of pathways such carotenoid biosynthetic pathway [Citation6,Citation7], accessory gene regulator (agr) [Citation8–10], and sortase enzymes that anchor surface proteins to the cell wall [Citation11–18].
Sortases are transpeptidases found in many Gram-positive bacteria [Citation14,Citation19,Citation20]. Among them, S. aureus sortase A (SrtA) is the archetypal sortase that cleaves secreted protein substrates between the threonine and glycine residues of the conserved C-terminal LPXTG sorting motif and links the newly released threonine residue to the pentaglycine cross-bridge of S. aureus peptidoglycan, thereby effectively immobilizing substrates into the cell wall [Citation14]. In S. aureus, 20 proteins with LPXTG sorting signals fulfill diverse functions such as iron acquisition from the host, bacterial adhesion, and immune evasion during the infectious process [Citation14]. SrtA plays a critical role in the pathogenesis of Gram-positive bacterial infections and a S. aureus srtA knockout mutant is unable to cause bacteraemia or sepsis in mice [Citation11,Citation14,Citation21].
SrtA is not required for bacterial viability in vitro and belongs to an exclusively bacterial family of enzymes, suggesting that specific SrtA inhibitors may have a limited selective pressure to promote the rise of resistance traits and are unlikely to exhibit particularly deleterious side effects [Citation16]. SrtA is thus considered an ideal target for anti-virulence drug development [Citation14–24]. The transpeptidase activity of SrtA has also been exploited in a range of industrial applications, including protein conjugation and targeted labelling of cell surface macromolecules [Citation25–32]. To date, S. aureus SrtA is the most understood of all the sortases and its molecular features (i.e. structures, functions, and mechanisms) have been extensively studied [Citation14,Citation20] subsequent to its discovery in 1999 [Citation11].
SrtA contain three conserved residues within its active site: His120, Cys184, and Arg197 [Citation33]. It is clear that Cys184 forms a covalent linkage to the sorting signal while the functions of His120 and Arg197 are controversial [Citation34]. Currently, the specificity and kinetic/catalytic mechanism of sortases remains an active area of research and a reverse protonation chemical mechanism of S. aureus SrtA has been proposed recently [Citation34]. In this mechanism, C184 thiolate is the active site nucleophile, H120 imidazolium is a general acid, and R197 is a transition state stabilizer [Citation34].
Although insight into the biological function, structure, specificity and mechanism of S. aureus SrtA is significant at present [Citation14,Citation20,Citation34], the mechanisms that may enable S. aureus to regulate the activity of SrtA have not been described. Here, we show that S. aureus SrtA is subject to post-translational regulation. We found that phosphorylation markedly decreased the enzyme activity of S. aureus SrtA. We also showed that SrtA is a key factor for the Stk1/Stp1 signalling pathway in S. aureus.
Results
SrtA is a substrate for Stk1 in vitro
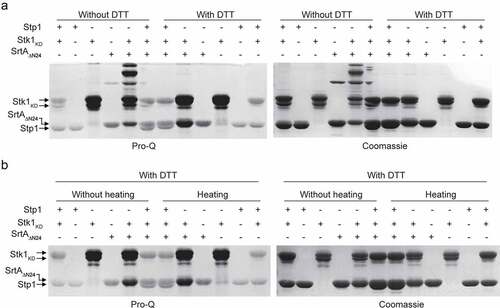
Eukaryote-type Ser/Thr protein kinases (STKs) and phosphatases (STPs) have emerged as critical signalling molecules in prokaryotes [Citation35,Citation36]. In S. aureus, Stk1 and Stp1 provide an additional level of regulation for a variety of biological functions, including cell wall biosynthesis, drug resistance and virulence regulation [Citation36–39]. To examine whether SrtA is a substrate for Stk1, the catalytic domain of S. aureus Stk1, termed Stk1KD, was cloned, expressed, and purified from E. coli. Using SDS-PAGE and autoradiography, we found that SrtAΔN24, a truncated tagged form of S. aureus SrtA [Citation40], was phosphorylated by Stk1KD in vitro (). Similar results were obtained using the Pro-Q Diamond and Coomassie blue stains with purified SrtAΔN59 (), another truncated S. aureus SrtA lacking the N-terminal 59 residues [Citation40]. In the presence of Stk1KD, the SrtAΔN59 band stained intensely with Pro-Q Diamond but not with Coomassie blue (). These results clearly suggest that SrtA is a substrate of Stk1 in vitro. Either phosphorylated SrtAΔN24 () or phosphorylated SrtAΔN59 () was dephosphorylated by Stp1, indicating that phosphorylated SrtA is a substrate of Stp1.
Figure 1. Reversible phosphorylation of SrtA and Co-IP experiments. (A) Recombinant SrtAΔN24 was incubated with the recombinant Stk1KD in the presence of [γ-32P]ATP. Stp1 proteins were added subsequent to the in vitro phosphorylation reaction in order to dephosphorylate SrtAΔN24 as indicated. Recombinant SrtAΔN24 and Stp1 have similar molecular weights (~27 KD) and migrate similarly in SDS-PAGE. (B) Reversible phosphorylation of SrtA by Stk1 and Stp1. (C) Phosphorylation of SrtAΔN24 in the context of a bacterial lysate. WT, cell lysates from wild-type Newman strain (harbouring pYJ335); Δstp1, cell lysates from a Δstp1 strain (harbouring pYJ335); stk1+++, cell lysates from S. auerus strain overexpressing the stk1 (Newman/pYJ335:stk1). (D and E) Dephosphorylation of recombinant SrtAΔN59 expressed and purified from either Δstp1 or WT Newman strain, band intensities from Pro-Q Diamond chemiluminescence were normalized against the corresponding coomassie brilliant blue stained band, which were set to 1.0 for purified recombinant SrtAΔN59 from the Δstp1 strain. In A-E, Reactions were stopped by addition of SDS – PAGE loading buffer (without DTT), samples were separated by SDS-PAGE, visualized by either autoradiography (32P) or pro-Q diamond stained gel (Pro-Q), and stained with coomassie blue. (F) Co-IP between Stk1 and SrtA. IP was performed with anti-Stk1 antibody followed by immunoblotting with anti-SrtA antibody (SrtA) and anti-Stk1 antibody (Stk1). The beads control was obtained in the absence of Stk1 antibody. (G) Co-IP between Stp1 and SrtA. IP was performed with anti-Stp1 antibody followed by immunoblotting with anti-SrtA antibody (SrtA) and anti-Stp1 antibody (Stp1). The beads control was obtained in the absence of Stp1 antibody. (H) Co-IP between SrtA and Stk1 or Stp1. IP was performed with anti-SrtA antibody followed by immunoblotting with anti-Stk1 antibody (Stk1), anti-Stp1 antibody (Stp1), and anti-SrtA antibody (SrtA). The beads control was obtained in the absence of SrtA antibody. In F-H, WT denotes WT S. aureus Newman strain, Δstp1 denotes stp1 deletion mutant, stk1-I denotes S. aureus Newman mutant with transposon insertion in stk1 gene, ΔsrtA denotes srtA deletion mutant.
![Figure 1. Reversible phosphorylation of SrtA and Co-IP experiments. (A) Recombinant SrtAΔN24 was incubated with the recombinant Stk1KD in the presence of [γ-32P]ATP. Stp1 proteins were added subsequent to the in vitro phosphorylation reaction in order to dephosphorylate SrtAΔN24 as indicated. Recombinant SrtAΔN24 and Stp1 have similar molecular weights (~27 KD) and migrate similarly in SDS-PAGE. (B) Reversible phosphorylation of SrtA by Stk1 and Stp1. (C) Phosphorylation of SrtAΔN24 in the context of a bacterial lysate. WT, cell lysates from wild-type Newman strain (harbouring pYJ335); Δstp1, cell lysates from a Δstp1 strain (harbouring pYJ335); stk1+++, cell lysates from S. auerus strain overexpressing the stk1 (Newman/pYJ335:stk1). (D and E) Dephosphorylation of recombinant SrtAΔN59 expressed and purified from either Δstp1 or WT Newman strain, band intensities from Pro-Q Diamond chemiluminescence were normalized against the corresponding coomassie brilliant blue stained band, which were set to 1.0 for purified recombinant SrtAΔN59 from the Δstp1 strain. In A-E, Reactions were stopped by addition of SDS – PAGE loading buffer (without DTT), samples were separated by SDS-PAGE, visualized by either autoradiography (32P) or pro-Q diamond stained gel (Pro-Q), and stained with coomassie blue. (F) Co-IP between Stk1 and SrtA. IP was performed with anti-Stk1 antibody followed by immunoblotting with anti-SrtA antibody (SrtA) and anti-Stk1 antibody (Stk1). The beads control was obtained in the absence of Stk1 antibody. (G) Co-IP between Stp1 and SrtA. IP was performed with anti-Stp1 antibody followed by immunoblotting with anti-SrtA antibody (SrtA) and anti-Stp1 antibody (Stp1). The beads control was obtained in the absence of Stp1 antibody. (H) Co-IP between SrtA and Stk1 or Stp1. IP was performed with anti-SrtA antibody followed by immunoblotting with anti-Stk1 antibody (Stk1), anti-Stp1 antibody (Stp1), and anti-SrtA antibody (SrtA). The beads control was obtained in the absence of SrtA antibody. In F-H, WT denotes WT S. aureus Newman strain, Δstp1 denotes stp1 deletion mutant, stk1-I denotes S. aureus Newman mutant with transposon insertion in stk1 gene, ΔsrtA denotes srtA deletion mutant.](/cms/asset/8299ad45-1ff7-40b9-aa3c-48800f38d16b/kvir_a_2171641_f0001_oc.jpg)
To further examine the roles of Stk1 and Stp1 in the phosphorylation of SrtA, we performed phosphorylation assays by incubating SrtAΔN24 proteins respectively with cell lysates of a wild-type (WT) Newman strain, a Δstp1 stain, and a Stk1-overproducing strain (stk1+++, the WT Newman strain carrying pYJ335:stk1). When cell lysates of either Δstp1 or stk1+++ strain was used, a higher phosphorylation level of SrtAΔN24 was observed compared to that obtained using the cell lysates of WT Newman strain (). Thus, both stp1 deletion and over-expression of stk1 facilitate the phosphorylation of SrtA in the context of a cell lysate. These results are also consistent with in vitro reversible phosphorylation of SrtA mediated by purified recombinant Stk1 and Stp1 ().
Phosphorylation of recombinant SrtA expressed in S. aureus
To probe the existence of phosphorylation of SrtA in vivo, we purified SrtAΔN59 from Δstp1 and WT S. aureus Newman strain respectively, and dephosphorylation assay was performed by incubating the SrtAΔN59 protein with Stp1. As shown in (), Stp1-treatment decreased the intensity of SrtAΔN59 band stained with Pro-Q Diamond but not with Coomassie blue, indicating that SrtA is phosphorylated in vivo. SrtAΔN59 proteins purified from the Δstp1 strain displayed higher intensity of Pro-Q Diamond stained band than those purified from the WT Newman strain (Coomassie staining as loading control) (). We also noted that Stp1-treatment decreases the phosphorylation level of SrtAΔN59 purified from the WT Newman strain (), but to a much lesser extent than it did for the SrtAΔN59 that had been purified from the Δstp1 strain (). Based on these data, we conclude that SrtA is very likely to be subject to reversible phosphorylation in vivo, and this effect appears to be mediated by at least Stk1 and Stp1. Supporting this view, Co-Immunoprecipitation (Co-IP) assays demonstrated the interactions between SrtA and Stk1, and between SrtA and Stp1 ().
SrtA contains multiple phosphorylation sites
To further verify the phosphorylation of SrtA and to identify the phosphorylation sites, we performed mass spectrometric characterization on the SrtA protein phosphorylated by the cell lysates of stk1+++ strain. Searching tandem mass spectrometry (MS/MS) spectra for neutral loss peaks of 80 daltons revealed the presence of at least 28 phosphorylation sites and that the SrtA protein can be phosphorylated simultaneously at multiple residues (, Fig. S1 A to E, and Table S3). These phosphorylation sites are mainly located in and around strands ß1, ß3 and ß4, strand ß6 and several amino acid residues close to the end of it, and strand ß7 of the crystal structure of SrtA () [Citation41]. The catalytic residues of S. aureus SrtA, Cys184, and His120 [Citation14], were subject to phosphorylation (, Fig. S1). Asp112, which is important for calcium binding [Citation42] and SrtA activity [Citation43], was also a potential phosphorylation site (, Table S3). These observations indicate that SrtA contains multiple phosphorylation sites.
Figure 2. Different phosphorylation events on SrtA. (A) Phosphorylation sites of SrtA revealed by mass spectrometry (MS/MS). Phosphorylation sites are highlighted with asterisk, the catalytic residues of SrtA (Cys184, His120, and Arg197) are highlighted with triangle, and some secondary structural elements are indicated according to the structure of S. aureus SrtA (PDB ID 1T2P). (B) A representative MS/MS spectrum of phosphorylation on the Cys184 of SrtA. The phosphor-peptide QLTLITpCDDYNEK was observed at m/z 818.3611 Da obtained after trypsin digestion of SrtA that has been phosphorylated by the bacterial cell lysates of stk1+++ strain. Individual fragments are labelled based on the b- or y-ion nomenclature. (C) In vitro phosphorylation of SrtAΔN24 and its variant by bacterial cell lysates of stk1+++ strain in the presence of [γ-32P]ATP. stk1+++, stk1-overexpressing strain (Newman/pYJ335:stk1). SrtAΔN24 C184A, variant of SrtAΔN24 harbouring a Cys to Ala substitution at C184. (D) Effect of DTT on the phosphorylation of SrtA that has been purified from Δstp1 strain. (E) Effect of AcP (25 mM) on the phosphorylation of SrtAΔN24 and ClpP in vitro. (F) Effect of sample heating or Stp1-treatment on the AcP-mediated phosphorylation of SrtAΔN24. (G) SrtAΔN59 C184A could be phosphorylated by acetyl phosphate (25 mM). (H) Phosphorylation of SrtAΔN24 C184A by Stk1. SrtAΔN24 and SrtAΔN24 C184A were respectively incubated with recombinant Stk1KD in the presence of [γ-32P]ATP; Stp1 was added subsequent to in vitro phosphorylation reaction in order to dephosphorylate SrtAΔN24 and SrtAΔN24 C184A. The reactions were stopped by addition of SDS – PAGE loading buffer (without DTT). Samples were separated by SDS-PAGE, visualized by autoradiography (32P) or Pro-Q diamond stained gel (Pro-Q), and stained with coomassie blue (Coomassie). recombinant SrtAΔN24 and Stp1 have similar molecular weights (~27 KD) and migrate similarly. SrtAΔN24 C184A, SrtAΔN24 variant harbouring a Cys to Ala substitution at C184.
![Figure 2. Different phosphorylation events on SrtA. (A) Phosphorylation sites of SrtA revealed by mass spectrometry (MS/MS). Phosphorylation sites are highlighted with asterisk, the catalytic residues of SrtA (Cys184, His120, and Arg197) are highlighted with triangle, and some secondary structural elements are indicated according to the structure of S. aureus SrtA (PDB ID 1T2P). (B) A representative MS/MS spectrum of phosphorylation on the Cys184 of SrtA. The phosphor-peptide QLTLITpCDDYNEK was observed at m/z 818.3611 Da obtained after trypsin digestion of SrtA that has been phosphorylated by the bacterial cell lysates of stk1+++ strain. Individual fragments are labelled based on the b- or y-ion nomenclature. (C) In vitro phosphorylation of SrtAΔN24 and its variant by bacterial cell lysates of stk1+++ strain in the presence of [γ-32P]ATP. stk1+++, stk1-overexpressing strain (Newman/pYJ335:stk1). SrtAΔN24 C184A, variant of SrtAΔN24 harbouring a Cys to Ala substitution at C184. (D) Effect of DTT on the phosphorylation of SrtA that has been purified from Δstp1 strain. (E) Effect of AcP (25 mM) on the phosphorylation of SrtAΔN24 and ClpP in vitro. (F) Effect of sample heating or Stp1-treatment on the AcP-mediated phosphorylation of SrtAΔN24. (G) SrtAΔN59 C184A could be phosphorylated by acetyl phosphate (25 mM). (H) Phosphorylation of SrtAΔN24 C184A by Stk1. SrtAΔN24 and SrtAΔN24 C184A were respectively incubated with recombinant Stk1KD in the presence of [γ-32P]ATP; Stp1 was added subsequent to in vitro phosphorylation reaction in order to dephosphorylate SrtAΔN24 and SrtAΔN24 C184A. The reactions were stopped by addition of SDS – PAGE loading buffer (without DTT). Samples were separated by SDS-PAGE, visualized by autoradiography (32P) or Pro-Q diamond stained gel (Pro-Q), and stained with coomassie blue (Coomassie). recombinant SrtAΔN24 and Stp1 have similar molecular weights (~27 KD) and migrate similarly. SrtAΔN24 C184A, SrtAΔN24 variant harbouring a Cys to Ala substitution at C184.](/cms/asset/adefc2b9-0a4b-44fc-847e-16ba43ead92b/kvir_a_2171641_f0002_oc.jpg)
Cysteine phosphorylation of SrtA
As aforementioned, Cys184 of S. aureus SrtA was subject to phosphorylation (). Because Cys184 is essential for the catalytic activity of SrtA and this residue is absolutely conserved and essential for sortase activity from Gram-positive bacteria [Citation14], we next sought to determine the role of Cys184 in the phosphorylation of SrtA. To this end, we analysed the phosphorylation levels of SrtAΔN24 and its mutant protein SrtAΔN24 C184A (in which Cys184 was replaced by an alanine residue) in the cell lysates of stk1+++ strain in the presence of [32P] ATP using autoradiography. Our results showed that phosphorylation occurred exclusively to the monomeric reduced forms of SrtAΔN24, but not to its dimeric oxidized forms and its mutant forms (SrtAΔN24 C184A) (). These observations suggest that Cys184 might be a key site responsible for the phosphorylation of SrtA in the context of a bacterial cell lysate.
Additionally, we observed that, in the presence of 1 mM dithiothreitol (DTT), phosphorylation of SrtAΔN24 by the cell lysates was almost abolished (). This result support the notion that Cys184 was very likely the major site of the observed phosphorylation (, lane 2) since cysteine phosphorylation is DTT-reversible [Citation36]. Using Pro-Q Diamond and Coomassie blue stains, we found that the phosphorylation of SrtAΔN59 that had been purified from Δstp1 stain displayed liability towards DTT (). Taken together, these biochemical data, along with the LC-MS/MS data (), suggest that Cys184 is a primary site for the phosphorylation of SrtA in the context of a bacterial cell lysate and cellular.
Different phosphorylation events on SrtA
Since several aspartic acid residues of S. aureus SrtA were subject to phosphorylation (, Fig. S1, and Table S3), we next sought to analyse the effect of acetyl phosphate (AcP) on the phosphorylation of SrtA, given that AcP can be used as a phosphodonor for the acceptor aspartic acid residue of some bacterial proteins such as response regulators [Citation44–49]. Using Pro-Q Diamond and Coomassie blue stains, we found that SrtAΔN24, but not Caseinolytic protease proteolytic subunit (ClpP) that serves as a control, can be efficiently phosphorylated by AcP (). We also observed that sample heating, which promotes extensive hydrolysis of phospho-Asp residues [Citation50], caused essentially loss of phosphorylation of SrtAΔN24 that has been treated by AcP (). These results indicate that certain aspartic acid residue of S. aureus SrtA may serve as acceptor for the phosphodonor AcP. Additionally, Stp1-treatment also decreased the phosphorylation level of SrtAΔN24 mediated by AcP (), indicating that Stp1 may possess aspartic acid dephosphorylation activity.
AcP was able to efficiently phosphorylate not only the SrtAΔN24 () but also the SrtAΔN59 C184A (). Additionally, the cysteine residue does not play an important role in the in vitro phosphorylation of SrtA by purified Stk1KD (). Thus, the Cys184 is unlikely a primary phosphorylation site of SrtA under these experimental conditions, although it did in the context of a bacterial cell lysate () and cellular (). Moreover, we observed that phosphorylated SrtAΔN24 that had been phosphorylated by purified Stk1KD was not susceptible to either DTT treatment () or sample heating (). In all, these data suggest that SrtA can contain different types of phosphorylation with distinct chemical properties including DTT-labile (), heat-labile (), DTT-stable (), and heat-stable (). These distinct chemical properties are probably due to phosphorylation occurring on different forms of amino acid residues, which is in line with the observation that various forms of amino acid residues of SrtA were phosphorylatable (, Fig. S1, and Table S3).
Figure 3. In vitro phosphorylation of SrtAΔN24. (A) Effect of DTT on Stk1-mediated phosphorylation of SrtAΔN24. The reaction was stopped by addition of SDS-PAGE loading buffer containing without or with DTT at a final concentration of 50 mM. samples were separated by SDS – PAGE, visualized by pro-Q diamond stained gel (left panel), and stained with Coomassie Blue (right panel). (B) Effect of sample heating on Stk1-mediated phosphorylation of SrtAΔN24. The reaction was stopped by addition of SDS – PAGE loading buffer (with DTT at a final concentration of 50 mM) without or with sample heating (95 °C, 10 min). samples were separated by SDS – PAGE, visualized by pro-Q diamond stained gel (left panel), and stained with coomassie blue (right panel).
Phosphorylation inhibits the enzymatic activity of SrtA
Cys184 is essential for the functions of SrtA [Citation14], we thus reasoned that its phosphorylation may abolish the enzyme activity of SrtA. We performed a fluorescence resonance energy transfer (FRET) assay with phosphorylated SrtAΔN24 (P~SrtAΔN24) that has been phosphorylated by the cell lysates of the stk1+++ strain, where cysteine residue is the primary phosphorylation site (). In this assay, Stp1 and DTT were respectively added to a similar reaction in order to dephosphorylate the P~SrtAΔN24. Our results showed that the treatment of purified P~SrtAΔN24 with Stp1 obviously increased its ability to cleave the abz-LPETG-dnp substrate (). DTT-treatment also gave rise to an obvious increase of SrtA enzyme activity (). Thus, cysteine phosphorylation appears to inhibit the enzyme activity of SrtA. However, it was a little unexpected that the purified P~SrtAΔN24 still exhibited enzyme activity () and the reasons for this are currently unknown. It may be due to the chemical liability of the S-linked phosphorylation of SrtA and/or the fact that the purified protein sample is a mixture of phosphorylated SrtAΔN24 with different phosphorylation events, except for the cysteine phosphorylation.
Figure 4. Phosphorylation inhibits the enzymatic activity of SrtA in vitro. (A) Effect of Stp1 and DTT on the enzyme activity of phosphorylated SrtAΔN24 (P~SrtAΔN24) in vitro. P~SrtAΔN24 was purified from E. coli-expressed SrtAΔN24 that has been phosphorylated by the cell lysates of Δstp1 strain. Stp1 and DTT were respectively added to the reaction mixture in order to dephosphorylate P~SrtAΔN24. (B) Effect of AcP-treatment on the in vitro enzyme activity of SrtAΔN24. Purified SrtAΔN24 expressed in E. coli was treated by AcP (25 mM) as described in materials and methods. Stp1 was added to the reaction mixture in order to dephosphorylate the AcP-mediated phosphorylation of SrtAΔN24. (C) Effect of Stk1KD and Stp1 on the enzyme activity of SrtAΔN24 in vitro. Recombinant Stk1KD was added to the reaction mixture in order to phosphorylate SrtAΔN24 while Stp1 was added in order to dephosphorylate SrtAΔN24. In (A) to (C), a fluorescently self-quenched peptide, Abz-LPETG-Dnp, was used to mimic the surface protein substrate, which contains a LPETG sorting signal. When Abz-LPETG-Dnp is cleaved by SrtA, the fluorophore Abz group is released from the quencher Dnp group, and an enhanced fluorescence signal was detected. RFI, relative fluorescence intensity.
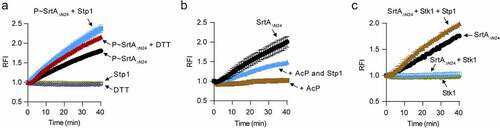
AcP – mediated phosphorylation also inhibited the enzyme activity of SrtA. As shown, AcP-treated SrtAΔN24 displayed almost no enzyme activity in our FRET assay while retreatment of the AcP-treated SrtAΔN24 with Stp1 obviously increased its ability to cleave the abz-LPETG-dnp substrate (). AcP might have a role in the regulation of SrtA-mediated anchoring of surface proteins in S. aureus. However, this hypothesis awaits further investigations. Nonetheless, these data clearly suggest that the inhibitory effect of AcP on the in vitro enzyme activity of SrtA is mediated at least in part by protein phosphorylation.
Additionally, we observed that the purified E. coli-expressed S. aureus SrtAΔN24 can cleave the abz-LPETG-dnp substrate efficiently () and as expected [Citation12] while it failed to do this in the presence of Stk1KD (). When Stp1 was subsequently added to the phosphorylation reaction mixture, the enzyme activity of SrtAΔN24 was restored (), indicating that Stk1-mediated phosphorylation inhibits the enzyme activity of SrtA. Based on these data, we conclude that the enzyme activity of SrtA is regulated by phosphorylation, at least in vitro.
Deletion of stp1 in S. aureus Newman produces srtA-deficient phenotypes
Deletion of stp1 caused an increase in the phosphorylation level of SrtA in S. aureus (), we thus reasoned that stp1 deletion may cause an inhibition of the function of SrtA because phosphorylation inhibited the in vitro enzymatic activity of SrtA (). Supporting this notion, we found that, like the srtA deletion mutant [Citation11,Citation14], Δstp1 strain exhibited severe defects in bacterial growth in CRPMI (an iron-restricted medium) supplemented with either haemoglobin () or haem () as the sole iron source. Δstp1 strain displayed slight growth defect only when FeSO4 was supplemented as the sole iron source, and as expected, all the testing S. aureus strains fail to grow in the iron-restricted CRPMI medium (). The introduction of plasmid p-srtA can alleviate the growth defects of Δstp1 strain in the CRPMI medium supplemented with either haemoglobin or haem the sole iron source ().
Figure 5. Overexpression of srtA alleviates the defects of stp1 deletion mutant. (A and B) Effect of constitutive expression of either stp1 or srtA on the bacterial growth of Δstp1 mutant in iron-restricted RPMI medium (Chelex 100 resin-treated RPMI) supplemented with either haemoglobin (2.5 μg/ml) or haem (2 μM) as the sole iron source. data shown represent the mean ± SD from triplicate experiments. (C) Growth curves for S. aureus in iron-restricted RPMI medium (Chelex 100 resin-treated RPMI) supplemented with or without FeSO4 (300 μM). Data shown represent the mean ± SD (n = three biological replicates). (D) Bacterial virulence of S. aureus Newman and its variants in mouse infection models. BALB/c female mice (n = 10) were retro-orbitally injected with 4 × 106 CFU S. aureus bacteria; S. aureus colonization in murine liver was measured after 5 days of infection; each symbol represents the value for an individual mouse; horizontal bars indicate observation means and dashed lines mark limits of detection; *p<0.05, ***p<0.001 (Mann–Whitney Test, two-tailed). S. aureus WT Newman harbours vector pYJ335 (Newman); stp1 deletion mutant harbours plasmid pYJ335 alone (Δstp1), pYJ335:stp1(Δstp1/p-stp1) or pYJ335:srtA (Δstp1/p-srtA); srtA deletion mutant (ΔsrtA) harbours plasmid pYJ335 alone (ΔsrtA) or (ΔsrtA/p-srtA). (E) A proposed mechanism of SrtA phosphorylation. SrtA may act as a mediator in a variety of signalling pathways in S. aureus through integrating diverse phosphorylation events.
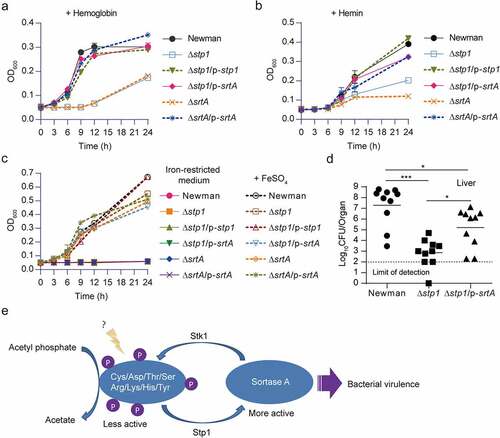
To examine the role of SrtA in Stp1-mediated signalling pathway during infections, we subjected the WT Newman, Δstp1, and Δstp1/p-srtA to a murine model of abscess formation and measured the bacterial survival in livers. As shown in (), the Δstp1 strain exhibited a significantly decreased colonization of liver compared to that of mice infected with the wild-type Newman (~4.5 log decrease in the CFU), whereas the introduction of plasmid p-srtA significantly increased the bacterial survival (~2.1 log increase in the CFU) (), indicating that ectopic expression of SrtA can suppress the virulence defect of Δstp1. Taken together, these results suggest that SrtA appears to be involved in the Stp1/Stk1 signalling pathway of S. aureus ().
Discussion
Here, we provide the first evidence that SrtA, an attractive anti-infective target for Gram-positive infections, is subject to phosphorylation that can inhibit its function (). Notably, the catalytic site residue of S. aureus SrtA [Citation14], Cys184, is a primary phosphorylation site in a cellular context ( B to 2D). Because Cys184 is essential for the enzyme activity and the function of SrtA [Citation14], it is thus difficult or impossible to interpret experiments involving mutations at this site unequivocally. However, consistent with previous reports that covalently modify this cysteine thiol abolished SrtA activity [Citation14], the enzymatic activity of P~SrtA that has been primarily phosphorylated at Cys184 is severely reduced when compared to that of Stp1-treated P~SrtA (). Moreover, the enzyme activity of P~SrtA was obviously increased in the presence of DTT (), which is in line with the observation that Cys-phosphorylation is DTT-labile () [Citation36]. S. aureus SrtA is highly resistant to oxidative inhibition and maintaining a high reduction potential of the Cys184 [Citation51]. It has also been reported that DTT fails to increase the enzymatic activity of SrtA even at a concentration of up to 10 mM [Citation52]. Given these, it is very likely that phosphorylation at Cys184 will introduce additional charge and conformational change, resulting in reduced enzyme activity of SrtA. We should, however, keep in mind that phosphorylation at other residue(s) may also contribute to the inhibition of the enzyme activity of SrtA ().
S. aureus SrtA protein contains multiple phosphorylatable amino acids (). Hence, in S. aureus the properties of SrtA (i.e. activity and specificity) may be shaped by different phosphorylation events, given that multisite phosphorylation is an important mechanism for fine-tuned regulation of protein function [Citation53,Citation54]. Meanwhile, phosphorylation occurring at various amino acid residues implies that SrtA may act as a mediator in a variety of signalling pathways in S. aureus through integrating diverse phosphorylation events. Indeed, we found that, at least in vitro, environmental conditions determine the phosphorylation events of SrtA (), and that phosphorylation inhibited its enzyme activity (). Unraveling these aspects of signal transduction mechanisms will open up the new horizon towards understanding of the regulation of surface display of staphylococcal proteins.
In this study, we used an established FRET-based assay to monitor the enzyme activity of SrtA in vitro [Citation52,Citation55,Citation56]. Although FRET has been extensively applied for probing the enzyme activity of SrtA, it is associated with several limitations [Citation17]. For example, as the assay depends on the measurements of fluorescence, the quencher product would probably interfere with the results due to the inner filter effect [Citation17]. Additionally, the presence of the cleaved product NH2-G-Dnp would decrease the fluorescence of Abz-LPET-OH when its concentration above 20 µM [Citation52]. Besides, the FRET-based assay can only provide information on the initial cleavage of LPXTG-containing peptides, but not the kinetic parameters associated with the transpeptidation step, since the fluorophore Abz group has already been departed from the Dnp quencher group after the completion of the first step [Citation56,Citation57].
In sum, to our best knowledge, this is the first report of the phosphorylation of a sortase enzyme in bacteria. Further studies on the detailed mechanism by which the phosphorylation events affect the enzyme activity of SrtA will provide new insights into how this protein is regulated in vivo and contribute to the development of potent SrtA inhibitors which aim to combat Gram-positive infections. Moreover, in this study, we determined that Stk1 and Stp1 play a role in the phosphorylation/de-phosphorylation of SrtA in vitro ( A to C) and these events are expected to happen in vivo ( E to H). Since phosphorylation events inhibit SrtA enzyme activity (), the inhibition of the Stp1 phosphatase might be a future pharmaceutical tool to reduce the activity of SrtA in S. aureus.
Materials and methods
Ethics statement
The animal study protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Public Health Clinical Center (Permit Number: 2013P201) and were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China [Citation11–14-]1988). The laboratory animal usage licence number is SYXK-HU-2010–0098, certificated by Shanghai Committee of Science and Technology. The animals were randomized but the investigators were not blinded to the experimental conditions.
Construction of plasmids
To construct the plasmid for the constitutive expression of srtA, a ca. 700 bp DNA covering 39 bp of srtA upstream region, the srtA gene, and a 40 bp downstream of srtA was amplified from S. aureus Newman genomic DNA with primers srtA-F and srtA-R (Table S2), and then cloned into pYJ335. The construct with srtA in the same orientation as the tetracycline-inducible xyl/tetO promoter was confirmed by DNA sequencing, yielding plasmid pYJ335:srtA.
To construct pYJ335:6his-srtAΔN59, a DNA fragment was amplified from pET28a:6his-srtAΔN59 with primers T7 promoter and T7 terminator (Table S2), and cloned into pYJ335. Primer pairs of pYJ335-F (Table S2) and T7 terminator were used to select the plasmid clones in which the genes are located downstream of the tetracycline-inducible xyl/tetO promoter.
To construct pET28a:6his-stk1KD, DNA fragment encoding the Stk1 kinase domain (Stk1KD) (residues 1–291) of S. aureus strain Newman was amplified by PCR with primers stk1-KD-F (with EcoRI site) and stk1-KD-R (with XhoI site). After digestion with EcoRI and XhoI, the PCR product was inserted into the pET28a vector and yielded pET28a:6his-stk1KD.
To construct pET28a:srtAΔN59, primer srtAΔN59-F and srtAΔN59-R (Table S2) were used. To introduce a single amino acid substitution (Cys to Ala substitution at C184) to srtA gene, primer pair srtAC184A-F/srtAC184A-R and plasmid DNA of either pET28a:srtAΔN24 or pET28a:srtAΔN59 were used as template. All the selected plasmid clones were sequenced to confirm that no additional mutations were introduced by PCR reactions.
Construction of S. aureus ΔsrtA mutant
The gene replacement vector pKOR1 was used to construct an in-frame, unmarked srtA deletion mutant (ΔsrtA). PCRs were performed to amplify flanking sequences of the intended deletion. The upstream fragment was amplified from S. aureus strain Newman genomic DNA using primers srtA-delete-up-F and srtA-delete-up-R (with EcoRI site), and the downstream fragment was amplified with primers srtA-delete-down-F (with EcoRI site) and srtA-delete-down-R. PCR products were digested with EcoRI, mixed together and ligated by T4 DNA ligase (New England Biolabs). The ligation product was amplified with primers srtA-delete-up-F and srtA-delete-down-R. Next, the PCR product was used for recombination with pKOR1, and the product was introduced to E. coli DH5α. The construction was sequenced to ensure that no additional mutations resulted. The resulting plasmid, pKOR1:ΔsrtA, was electroporated to S. aureus RN4220, and subsequently into S. aureus Newman. The allelic replacement was performed as described previously [Citation36] and the deletion of srtA was further confirmed by PCR and DNA sequencing.
Recombinant protein purification from E. coli
Transformed bacteria were sub-cultured into 1000 ml LB medium supplemented with 50 μg/ml kanamycin at 37°C to an optical density of 0.5 at 600 nm. After lowering the temperature to 16°C (for Stp1 and SrtA expression) or 20°C (for Stk1KD expression), 1 mM isopropyl-thiogalactopyranoside (IPTG) was added in order to induce the expression of protein and the bacteria were harvested by centrifugation after an overnight of shaking (250 rpm of aeration).
For the purification of 6His-Stk1KD, the pellets were resuspended in 30 ml buffer A [20 mM HEPES, pH 7.4; 150 mM NaCl, 10 mM imidazole, and 1 mM phenylmethanesulfonyl fluoride (PMSF)] and sonicated. The lysate was centrifuged at 15,000 g for 30 min, and the supernatant was loaded onto a HisTrap HP 5 ml column (GE Healthcare), which was then equilibrated with buffer A. The 6His-Stk1KD was eluted with a linear gradient of 10–500 mM imidazole with buffer B [20 mM HEPES, pH 7.4; 150 mM NaCl, 500 mM imidazole, and 1 mM phenylmethanesulfonyl fluoride (PMSF)]. Fractions enriched for 6His-Stk1KD were pooled and concentrated. The proteins were further purified by gel filtration (Superdex 75, GE Healthcare) with buffer C [20 mM HEPES, pH 7.4; 150 mM NaCl], stored at −80°C at a concentration of approximately 1 mg/ml in buffer D [10 mM HEPES, pH 7.4; 75 mM NaCl, 30% (wt/vol) glycerol].
Purification of Stp1 protein was performed as previously described [Citation36]. For the purification of 6His-SrtA and its variants, buffer I [50 mM Tris-HCl, pH 7.5; 200 mM NaCl, 5 mM 2-mercaptoethanol, 40 mM imidazole] and buffer J [50 mM Tris-HCl, pH 7.5; 200 mM NaCl, 5 mM 2-mercaptoethanol, 300 mM imidazole] were used, and buffer K [20 mM Tris-HCl, pH 7.5, 50 mM NaCl] was used as gel filtration buffer. The purified proteins were stored in buffer L [10 mM Tris-HCl, pH 7.5, 25 mM NaCl, 30% (wt/vol) glycerol] at −80°C at a concentration of approximately 1 mg/ml until use.
Protein phosphorylation by purified 6his-Stk1KD
For the detection of phosphoproteins with autoradiography, in vitro kinase assay were performed by incubating 0.5 μg 6His-Stk1KD without or with 4 μg of the desired proteins (6His-SrtAΔN24 or 6His-SrtAΔN24 C184A, as indicated) in phosphorylation reaction buffer [50 mM Tris-HCl, pH 7.5; 0.1 mM EDTA, 1 mM MnCl2, 5 mM MgCl2] in the presence of 10 μCi [γ-32P]ATP at room temperature for 30 min. Likewise, for dephosphorylation assay, reaction was incubated with 0.5 μg Stp1 for an additional 30 min at room temperature. Reactions were performed in volumes of 20 μl, and an equal volume of protein storage buffer was added as needed in order to maintain a constant reaction condition. Reactions were stopped by the addition of 5 μl 5 × SDS loading buffer (without DTT), and the samples were resolved on a 12% SDS-PAGE, visualized by autoradiography.
Similarly, for the detection of phosphoproteins by Pro-Q Diamond stain, the in vitro kinase assay was performed by incubating 2 μg 6His-Stk1KD without or with 4 μg of the desired proteins in phosphorylation reaction buffer supplemented with 1 mM ATP at room temperature for 30 min. Likewise, the dephosphorylation assay was performed by incubating the reactions with 1 μg Stp1 for an additional 30 min at room temperature. Reaction was stopped by the addition of 5 μl 5 × SDS loading buffer (with or without DTT, as indicated). Samples were resolved on a 12% SDS-PAGE. Following electrophoresis, proteins were immersed 30 min in fixing solution [50% methanol, 10% acetic acid]. The gel was subsequently washed in distilled water and treated for 90 min with Pro-Q Diamond Phosphoprotein Gel Stain. After destaining with destain buffer [20% acetonitrile, 50 mM sodium acetate, pH 4.0], phosphorylated species were visualized by using Tanon-5200 multi.
All the gels were subsequently stained with Coomassie blue to ensure the quantity of the loaded proteins.
Protein phosphorylation by bacterial cell lysates
S. aureus overnight cultures were diluted 100-fold in fresh TSB medium, and grown in a 250-ml flask with a flask volume to medium volume ratio of 5:1, shaking with 250 rpm at 37°C for 6 h. 10 ml cultures were harvested by centrifugation, washed once and suspended in 1 ml of phosphorylation buffer [50 mM Tris-HCl, pH 7.5; 0.1 mM EDTA, 1 mM MnCl2, 5 mM MgCl2] supplied with 1 μl protease inhibitor cocktail (Thermo) and 1 μl phosphatase inhibitor cocktail 2 (Sigma). The mixture was homogenized by mechanical disruption (Fast Prep FP2400 instrument; Qbiogene) and then removed debris by centrifuging and filtrating through 0.45 μm filter.
For phosphorylation assay, 30 μg cell lysates were incubated with or without the desired 6His-proteins (~2 μg) in a phosphorylation buffer in the presence of 10 μCi of [γ-32P] ATP. Reactions were performed in volumes of 20 μl at room temperature for 30 min, and stopped by adding 5 μl 5 × SDS loading buffer (with or without DTT, as indicated). The phosphorylation signal was visualized by autoradiography and the gel was subsequently stained with Coomassie blue to ensure the quantity of the loaded proteins.
Protein phosphorylation by acetyl phosphate
Recombinant SrtA proteins (~4 µg) purified from E. coli were incubated with or without lithium potassium acetyl phosphate (AcP) (a final concentration of 25 mM) in a phosphorylation reaction buffer. Reactions were performed in volumes of 20 μl at 37°C for 1 h. 6his-tagged Pseudomonas aeruginosa ClpP protein (6His-ClpP) [Citation58], which contains 13 aspartic acid residues, was included as a recombinant protein controls, where appropriate. Reactions were stopped by adding 5 μl 5 × SDS loading buffer (with 100 mM DTT), and the samples were resolved on SDS-PAGE, and visualized by Pro-Q Diamond stain, subsequently stained with Coomassie blue to ensure the quantity of the loaded proteins. The signal intensity was determined by densitometric analysis using ImageQuant software.
For the dephosphorylation assay, recombinant proteins that have treated by AcP were loaded to a desalting spin column (BIORAD Bio-Spin® 6 column) for buffer exchange with 10 mM Tris buffer (10 mM Tris-HCl, pH 7.4) containing 0.02% sodium azide. After that, AcP-treated proteins (~4 µg) were incubated with or without Stp1 (~1 µg) at 37°C for 30 min, or subject to heating (95°C for 10 min). An equal volume of protein storage buffer was added as needed in order to maintain a constant reaction condition and reactions were performed in a phosphorylation reaction buffer in volumes of 20 μl.
Dephosphorylation of 6his-SrtAΔN59 purified from S. aureus
Overnight culture of S. aureus was diluted 1:100 in fresh 1,000 ml TSB (containing 10 μg/ml chloramphenicol, 10 μg/ml erythromycin, and 0.1 μg/ml anhydrotetracycline) and inculcated at 37°C with for 6 h with shaking (250 rpm of aeration). Cultures were harvested by centrifugation, washed once and suspended in 40 ml of buffer M [20 mM Na3PO4, pH 7.5; 200 mM NaCl] supplied with 4 μl protease inhibitor cocktail and 4 μl phosphatase inhibitor cocktail 2. The mixture was homogenized by mechanical disruption (Fast Prep FP2400 instrument; Qbiogene) and then removed debris by centrifuging and filtrating through 0.45 μm filter. 6His-SrtAΔN59 was enriched with Ni-NTA beads and eluted with buffer N [20 mM Na3PO4, pH 7.5; 200 mM NaCl, 300 mM imidazole]. The eluted proteins were concentrated to about 1 mg/ml using Amicon Ultra centrifugal filter.
For protein dephosphorylation, 6His-SrtAΔN59 proteins (~4 μg) were incubated with Stp1 (~1 μg) or DTT in a phosphorylation buffer for final volume of 20 μl. In the control reaction (6His-SrtAΔN59 alone), an equal volume of protein storage buffer for Stp1 was added. The reaction mixtures were maintained at room temperature for 30 min and then were stopped by adding 5 μl 5 × SDS loading buffer (without DTT). The samples were resolved on SDS-PAGE, and visualized by Pro-Q Diamond stain, subsequently stained with Coomassie blue to ensure the quantity of the loaded proteins. The signal intensity was determined by densitometric analysis using ImageQuant software.
LC-MS/MS identification of phosphorylation sites on SrtA
6His-SrtAΔN59 (~60 μg) purified from E. coli was added into cell free extracts (~300 µg) of stk1+++ (Newman/pYJ335:stk1) and incubated with 1 mM ATP for 30 min at room temperature. The trypsin digested sample was analysed using an LTQ-Orbitrap Elite or Q exactive mass spectrometer using a nanospray source. After phosphopeptide enrichment by Fe-NTA spin column, 50 µg sample was loaded onto the MudPIT column. Full MS spectra (from m/z 300–1800) were acquired by the precursor ion scan using the Orbitrap analyser with resolution R = 60, 000 at m/z 400, followed by 20 MS/MS events. The twenty most intense ions were sequentially isolated and subjected to CID or HCD for fragmentation.
Protein identification analysis was done with Integrated Proteomics Pipeline (IP2, Integrated Proteomics Applications, Inc. San Diego, CA. http://www.integratedproteomics.com) using ProLuCID/Sequest, DTASelect2. Tandem mass spectra were extracted into ms2 files from raw files using RawExtract 1.9.9 (http://fields.scripps.edu/downloads.php) and were searched against Uniprot Staphylococcus aureus Newman local database. Database search criteria are as follows: mass tolerances for precursor ions were set at 12 ppm and for MS/MS were set at 600 ppm. Cysteine, serine, threonine, tyrosine, histidine, arginine, lysine and aspartic phosphorylation were considered as variable modifications and we require one peptide per protein and at least one tryptic terminus for each peptide identification.
Co-immunoprecipitation
Overnight cultures of S. aureus were diluted 1:100 and grown in TSB in a 250-ml flask with a tube volume-to-medium volume ratio of 5:1, shaking with 250 rpm at 37°C for 6 h. 10 ml cultures were harvested by centrifugation, washed once and suspended in 1 ml of lysis buffer provided in the CO-IP kit (Pierce) supplied with 1 μl protease inhibitor cocktail (Thermo). The mixture was homogenized by mechanical disruption (Fast Prep FP2400 instrument; Qbiogene) and then the debris was removed by centrifuging and filtrating through 0.45 μm filters. 500 µl clarified lysates were incubated with the resins which immobilized with specific antibodies or not for 12 h at 4°C. The bound materials were washed and eluted following the manufacturer’s recommendation. Total samples (clarified lysates) and the immunocomplexes eluted from the resins were analysed by western blot with specific antibodies.
Western blot analysis
15 μl of 5 × SDS loading buffer [250 mM Tris-HCl, pH 6.8; 10% SDS; 0.5% bromophenol blue; 50% glycerol; 100 mM DTT] was added into 60 μl eluted immunocomplexes and then heated at 100°C for 10 min. SDS-PAGE was carried out subsequently. Thereafter, proteins were transferred to a PVDF (Bio-Rad) membrane at 350 mA for 60 min. The membrane was incubated with the primary antibody in 10 ml of 5% (wt/vol) skim milk at 4°C overnight following the blocking step [10 ml of 5% (wt/vol) skim milk] at room temperature for 2 h, and then washed three times at room temperature for 30 min in TBST buffer [10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.04% Tween 20]. Then, membranes were incubated with the secondary antibody for 2 h at room temperature and washed three times for 30 min in TBST. The chemiluminescent detection reaction was performed and detected by Tanon-5200 multi. The following primary antibodies were used: anti-Sortase A antibody (Abcam, #ab13959) with a 1:3000 dilution, anti-Stk1 polyclonal antibody (prepared by immunizing rabbits with the purified 6His-Stk1KD, Shanghai Immune Biotech CO., Ltd) with a 1:1000 dilution, anti-Stp1 polyclonal antibody (prepared by immunizing rabbits with the purified Stp1 protein, Shanghai Immune Biotech CO., Ltd) with a 1:500 dilution. For detection of the primary antibodies, anti-rabbit IgG (GE Healthcare, #NA934) with a 1:5000 dilution was used.
Enrichment of phosphorylated SrtA
6His-SrtAΔN24 proteins (~15 mg) purified from E. coli was incubated with the cell lysates (~75 mg) of S. aureus Δstp1 strain in the presence of 1 mM ATP in phosphorylation buffer. Reactions were performed in volumes of 40 ml at room temperature for 30 min. 6His-SrtAΔN24 was purified with HisTrap HP 5 ml column and the phosphoprotein purification kit (Qiagen) was used to further enrichment of the phosphorylated forms of 6His-SrtAΔN24 (P ~ 6His-SrtAΔN24) following the manufacturer’s instruction. The enriched protein was concentrated to 1 mg/ml in phosphoprotein elution buffer [2.92 g NaCl, 7.42 g K2HPO4, 1.15 g KH2PO4, pH 7.5, ddH2O to 1,000 ml].
Enzymatic activity of SrtA
A FRET assay was used to monitor the in vitro activity of SrtA as described previously using LPETG fluorescent peptides (abz-LPETG-dnp) as a substrate [Citation52]. In a typical FRET assay, a 100 μl reaction mixture contained 3 μM LPETG fluorescent peptides (abz-LPETG-dnp), 4 μg 6His-SrtAΔN24 in assay buffer [50 mM Tris-HCl, pH 7.5; 150 mM NaCl, 5 mM CaCl2]. An equal volume of protein storage buffer (and/or phosphorylation buffer) was added as needed in order to maintain constant reaction conditions.
Briefly, for the dephosphorylation of 6His-SrtAΔN24 that has been phosphorylated by the cell lysates of S. aureus Δstp1, 12 µg phosphorylated proteins were incubated with either Stp1 (~3 µg) or DTT (final concentration of 1 mM) at 37°C for 30 min in phosphorylation buffer in a total volume of 60 µl and after that, 20 μl sample mixture is used for subsequent FRET assay. For the dephosphorylation of 6His-SrtAΔN24 that has been phosphorylated by AcP, AcP-terated 6His-SrtAΔN24 (12 µg) was incubated with 3 µg Stp1 in phosphorylation buffer in a total volume of 60 µl and incubated at 37°C for 30 min, and 20 μl sample mixture is then used for subsequent FRET assay.
For the phosphorylation of 6His-SrtAΔN24 by Stk1, in vitro kinase assay was performed in 60 μl of phosphorylation buffer containing 6His-SrtAΔN24 (~12 μg) and 6His-Stk1KD (~6 μg) in the presence of 1 mM ATP at 37°C for 30 min. Likewise, for the dephosphorylation assay, reaction was incubated with 3 μg Stp1 for an additional 30 min at room temperature. 20 μl sample mixture is then used for subsequent FRET assay, respectively.
Reaction sample was distributed in 96-well black plates (PerkinElmer), and the increase in fluorescence intensity was monitored every 1 min for 40 min using Synergy 2 (Biotek) plate reader at an excitation wavelength of 360 nm and emission wavelength of 460 nm. Triplicate measurements were taken for each sample, and the data are reported as mean.
Bacterial growth curves
The S. aureus cultures were grown overnight in RPMI supplied with 1% (wt/vol) casamino acids and 500 μM 2,2-dipyridyl (DIP) to generate iron-restricted condition. The cells were washed twice in CRPMI (Chelex treated RPMI) containing 500 μM DIP, and normalized to OD600 to 5.0. Subcultured 10 μl bacteria suspension into 1 ml CRPMI containing 25 μM ZnCl2, 25 μM MnCl2, 1 mM MgCl2, 100 μM CaCl2, 0.5 mM DIP, and grown in 15 ml screw-cap conical tube at 37°C with aeration. 2.5 μg/ml (wt/vol) haemoglobin (TCI, H1293), 2 μM haem (Alfa Aesar, A18518) or 300 μM FeSO4 was supplemented (at a final concentration) as indicated and OD600 was monitored in every 3 h or 12 h by removed 50 μl culture mixed with 150 μl PBS in 96-well plates. The data are reported as mean ± SD.
Mouse infection models
S. aureus overnight cultures were diluted 1:100 and grown in fresh TSB in a 250-ml flask with a tube volume-to-medium volume ratio of 5:1, shaking with 250 rpm at 37°C for 3 h. Bacteria were harvested and washed twice with ice-cold PBS. The CFU per millilitre was determined before mice were inoculated. Mouse infection models were carried out as described previously [Citation7,Citation59]. 6 to 8-week-old female BALB/c mice were obtained from SIPPR-BK Lab Animal ltd (http://www.slarc.org.cn) and housed under specified pathogen-free conditions.
Mice were anaesthetized with pentobarbital sodium (intra-peritoneal injection, 80 mg/kg) and were challenged with either 4 × 106 CFU of S. aureus Newman and its variants. Animals were euthanized 5 days post infection. Livers were aseptically removed and homogenized in PBS in order to obtain single-cell suspensions, and serial dilutions of each organ were plated on TSA plates for the enumeration of CFU. In this experiment, mice were also received intra-peritoneal injections every 24 h with a total dose of 40 mg/kg erythromycin hydrochloride (in order to maintain the plasmid) and 10 μg/kg anhydrotetracycline (in order to induce srtA expression) dissolved in sterile double-distilled water (ddH2O) for 120 h (5 d). The statistical significance was determined by the Mann–Whitney Test (two-tailed).
Author contributions
L.L. and F.C conceived and initiated the study; H.D. performed most of the experiments; C. P. performed MS analysis; F.C, H.D., Y.W., R. C., H.P., H.L and L.L. analysed the data; L.L. wrote the paper with inputs from F.C., H.D. and H. L. All authors contributed to interpretation of data, and read and approved the final manuscript.
Supplemental Material
Download MS Word (2.6 MB)Acknowledgments
This work was supported by grants from National Natural Science Foundation (NSFC) (grant nos. 32270184 and 31870127). This study was also funded by the Science and Technology Commission of Shanghai Municipality (grant no. 19JC1416400) and State Key Laboratory of Drug Research (SIMM2003ZZ-03 and SIMM2205KF-07).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2023.2171641
Additional information
Funding
References
- Fisher JF, Mobashery S. Beta-lactams against the fortress of the gram-positive staphylococcus aureus bacterium. Chem Rev. 2021;121(6):3412–16.
- Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9(2):117–128.
- Cegelski L, Marshall GR, Eldridge GR, et al. The biology and future prospects of antivirulence therapies. Nature rev Microbiol. 2008;6(1):17–27.
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–548.
- Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of staphylococcus aureus. Virulence. 2021;12(1):547–569.
- Liu CI, Liu GY, Song Y, et al. A cholesterol biosynthesis inhibitor blocks staphylococcus aureus virulence. Science. 2008;319(5868):1391–1394. DOI:10.1126/science.1153018
- Chen F, Di H, Wang Y, et al. Small-molecule targeting of a diapophytoene desaturase inhibits S. aureus virulence. Nat Chem Biol. 2016;12(3):174–179. DOI:10.1038/nchembio.2003
- Balaban N, Goldkorn T, Nhan Rt et al. Autoinducer of virulence as a target for vaccine and therapy against Staphylococcus aureus. Science. 1998;280(5362):438–440. DOI:10.1126/science.280.5362.438
- Lyon GJ, Mayville P, Muir TW, et al. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc Natl Acad Sci. 2000; 97(24):13330–13335.
- Sully EK, Malachowa N, Elmore BO, et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLOS Pathogens. 2014;10(6):e1004174. DOI:10.1371/journal.ppat.1004174
- Mazmanian SK, Liu G, Ton-That H, et al. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763.
- Zhang J, Liu H, Zhu K, et al. Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase. Proc Natl Acad Sci. 2014; 111(37):13517–13522.
- Cossart P, Jonquieres R. Sortase, a universal target for therapeutic agents against gram-positive bacteria?. Proc Natl Acad Sci. 2000; 97(10):5013–5015.
- Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60(1):128–141.
- Cascioferro S, Totsika M, Schillaci D. Sortase A: an ideal target for anti-virulence drug development. Microbial Pathogenesis. 2014;77:105–112.
- Alharthi S, Alavi SE, Moyle PM, et al. Sortase a (SrtA) inhibitors as an alternative treatment for superbug infections. Drug Discov Today. 2021;26(9):2164–2172.
- Sapra R, Rajora AK, Kumar P, et al. Chemical biology of sortase a inhibition: a gateway to anti-infective therapeutic agents. J Med Chem. 2021;64(18):13097–13130. DOI:10.1021/acs.jmedchem.1c00386
- Cascioferro S, Raffa D, Maggio B, et al. Sortase a inhibitors: Recent advances and future perspectives. J Med Chem. 2015;58(23):9108–9123. DOI:10.1021/acs.jmedchem.5b00779
- Spirig T, Weiner EM, Clubb RT. Sortase enzymes in gram-positive bacteria. Mol Microbiol. 2011;82(5):1044–1059.
- Bradshaw WJ, Davies AH, Chambers CJ, et al. Molecular features of the sortase enzyme family. FEBS J. 2015;282(11):2097–2114. DOI:10.1111/febs.13288
- Mazmanian SK, Liu G, Jensen ER, et al. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci. 2000;97(10):5510–5515.
- Wang L, Jing S, Qu H, et al. Orientin mediates protection against MRSA-induced pneumonia by inhibiting sortase a. Virulence. 2021;12(1):2149–2161. DOI:10.1080/21505594.2021.1962138
- Su X, Yu H, Wang X, et al. Cyanidin chloride protects mice from methicillin-resistant Staphylococcus aureus-induced pneumonia by targeting sortase a. Virulence. 2022;13(1):1434–1445. DOI:10.1080/21505594.2022.2112831
- Guan XN, Zhang T, Yang T, et al. Covalent sortase a inhibitor ML346 prevents Staphylococcus aureus infection of galleria mellonella. RSC Med Chem. 2022;13(2):138–149. DOI:10.1039/D1MD00316J
- Popp MW, Antos JM, Grotenbreg GM, et al. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3(11):707–708.
- Dorr BM, Ham HO, An C, et al. Reprogramming the specificity of sortase enzymes. Proc Natl Acad Sci. 2014;111(37):13343–13348.
- Ham HO, Qu Z, Haller CA, et al. In situ regeneration of bioactive coatings enabled by an evolved Staphylococcus aureus sortase a. Nat Commun. 2016;7(1):11140. DOI:10.1038/ncomms11140
- Wagner K, Kwakkenbos Mj, Claassen Yb, et al. Bispecific antibody generated with sortase and click chemistry has broad antiinfluenza virus activity. Proc Natl Acad Sci. 2014;111(47):16820–16825.
- Swee LK, Guimaraes Cp, Sehrawat S, et al. Sortase-mediated modification of alphaDEC205 affords optimization of antigen presentation and immunization against a set of viral epitopes. Proc Natl Acad Sci. 2013;110(4):1428–1433.
- Ge Y, Chen L, Liu S, et al. Enzyme-mediated intercellular proximity labeling for detecting cell–cell interactions. J Am Chem Soc. 2019;141(5):1833–1837. DOI:10.1021/jacs.8b10286
- Yamaguchi S, Ikeda R, Umeda Y, et al. Chemoenzymatic labeling to visualize intercellular contacts using lipidated sortaseA. Chembiochem. 2022;23(21):e202200474. DOI:10.1002/cbic.202200474
- Shen D, Bai Y, Liu Y. Chemical biology toolbox to visualize protein aggregation in live cells. Chembiochem. 2022;23(4):e202100443.
- Frankel BA, Tong Y, Bentley ML, et al. Mutational analysis of active site residues in the Staphylococcus aureus transpeptidase SrtA. Biochemistry. 2007;46(24):7269–7278.
- Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers. 2010;94(4):385–396.
- Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75(1):192–212.
- Sun F, Ding Y, Ji Q, et al. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci. 2012;109(38):15461–15466.
- Cheung A, Duclos B. Stp1 and Stk1: the Yin and Yang of vancomycin sensitivity and virulence in vancomycin-intermediate Staphylococcus aureus strains. J Infect Dis. 2012;205(11):1625–1627.
- Debarbouille M, Dramsi S, Dussurget O, et al. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol. 2009;191(13):4070–4081. DOI:10.1128/JB.01813-08
- Prust N, van der Laarse S, van den Toorn H, et al. In-depth characterization of the staphylococcus aureus phosphoproteome reveals new targets of Stk1. Mol Cell Proteomics. 2021;20:100034.
- Nitulescu G, Margina D, Zanfirescu A, et al. Targeting bacterial sortases in search of anti-virulence therapies with low risk of resistance development. Pharmaceuticals (Basel). 2021;14(5):415.
- Zong Y, Bice TW, Ton-That H, et al. Crystal structures of Staphylococcus aureus sortase a and its substrate complex. J Biol Chem. 2004;279(30):31383–31389.
- Ilangovan U, Ton-That H, Iwahara J, et al. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci. 2001;98(11):6056–6061.
- Naik MT, Suree N, Ilangovan U, et al. Staphylococcus aureus Sortase a transpeptidase. calcium promotes sorting signal binding by altering the mobility and structure of an active site loop. J Biol Chem. 2006;281(3):1817–1826. DOI:10.1074/jbc.M506123200
- Kenney LJ, Bauer MD, Silhavy TJ. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc Natl Acad Sci. 1995;92(19):8866–8870.
- McCleary WR, Stock JB. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269(50):31567–31572.
- Cao Q, Wang Y, Chen F, et al. A novel signal transduction pathway that modulates rhl quorum sensing and bacterial virulence in pseudomonas aeruginosa. PLOS Pathogens. 2014;10(8):e1004340. DOI:10.1371/journal.ppat.1004340
- Yu L, Cao Q, Chen W, et al. A novel copper-sensing two-component system for inducing Dsb gene expression in bacteria. Sci Bull. 2022;67(2):198–212. DOI:10.1016/j.scib.2021.03.003
- Cao Q, Yang N, Wang Y, et al. Mutation-induced remodeling of the BfmRS two-component system in pseudomonas aeruginosa clinical isolates. Sci Signal. 2020;13(656):eaaz1529. DOI:10.1126/scisignal.aaz1529
- Xu C, Cao Q, Lan L. Glucose-binding of periplasmic protein GltB activates GtrS-GltR two-component system in pseudomonas aeruginosa. Microorganisms. 2021;9(2):447.
- Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using phos-tag-based reagents. Anal Biochem. 2008;376(1):73–82.
- Melvin JA, Murphy CF, Dubois LG, et al. Staphylococcus aureus sortase a contributes to the trojan horse mechanism of immune defense evasion with its intrinsic resistance to Cys184 oxidation. Biochemistry. 2011;50(35):7591–7599. DOI:10.1021/bi200844h
- Huang X, Aulabaugh A, Ding W, et al. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry. 2003;42(38):11307–11315. DOI:10.1021/bi034391g
- Salazar C, Hofer T. Multisite protein phosphorylation–from molecular mechanisms to kinetic models. FEBS J. 2009;276(12):3177–3198.
- Bah A, Vernon RM, Siddiqui Z, et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519(7541):106–109. DOI:10.1038/nature13999
- Ton-That H, Mazmanian SK, Alksne L, et al. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem. 2002;277(9):7447–7452.
- Ton-That H, Mazmanian SK, Faull KF, et al. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275(13):9876–9881.
- Ton-That H, Liu G, Mazmanian SK, et al. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci. 1999;96(22):12424–12429.
- Yang N, Cao Q, Hu S, et al. Alteration of protein homeostasis mediates the interaction of pseudo-monas aeruginosa with Staphylococcus aureus. Mol Microbiol. 2020;114(3):423–442. DOI:10.1111/mmi.14519
- Ding Y, Liu X, Chen F, et al. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci. 2014;111(46):E4981–4990.

