Abstract
Aim: This study aims to investigate the passive diffusion of protein kinase inhibitors through the blood–brain barrier (BBB) and to develop a model for their permeability prediction. Materials & methods: We used the parallel artificial membrane permeability assay to obtain logPe values of each of 34 compounds and calculated descriptors for these structures to perform quantitative structure–property relationship modeling, creating different regression models. Results: The logPe values have been calculated for all 34 compounds. Support vector machine regression was considered the most reliable, and CATS2D_09_DA, CATS2D_04_AA, B04[N-S] and F07[C-N] descriptors were identified as the most influential to passive BBB permeability. Conclusion: The quantitative structure–property relationship-support vector machine regression model that has been generated can serve as an efficient method for preliminary screening of BBB permeability of new analogs.
Graphical abstract
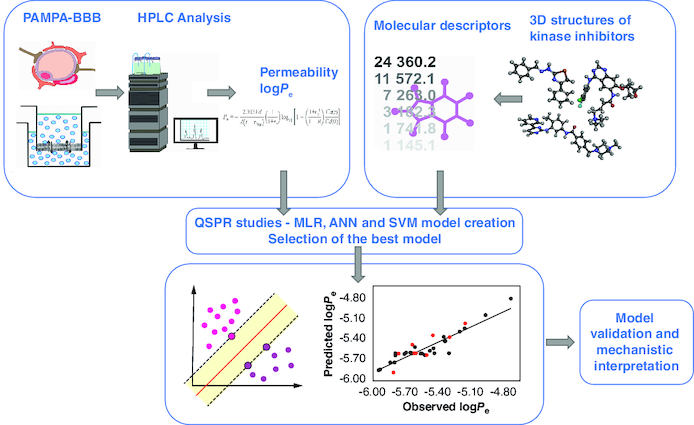
The parallel artificial membrane permeability assay in vitro technique was used to predict the ability of diverse kinase inhibitors and related compounds to penetrate the blood–brain barrier (BBB).
The experimentally determined permeability values (logPe) and the calculated molecular descriptors were analyzed using three different statistical and mathematical methods - multiple linear regression (MLR), artificial neural network (ANN) and support vector machine (SVM) to develop linear and nonlinear quantitative structure–property relationship (QSPR) models which can reliably predict BBB permeability.
To evaluate the predictive ability of the models, they were subjected to extensive statistical validation. The results showed that the SVM model has good predictive power based on the validation parameters, making it an effective tool for predicting the permeability of new analogs within the defined applicability domain.
It was found that the descriptors CATS2D_09_ DA, CATS2D_04_AA, B04[N-S], and F07[C-N] have the greatest influence on the logPe values. The integration of this technique with in silico computational methods provided a reliable, rapid and cost-effective methodology for assessing brain penetration.
The structural diversity of the compounds in the dataset and good predictive power of the created model indicated the potential utility of these methods for assessing permeability through the BBB of related kinase inhibitor candidates targeting CNS.
Author contributions
M Jovanović: conceptualization, methodology, software, writing – original draft. M Radan: methodology, supervision, writing – review & editing. M Čarapić: formal analysis, writing – original draft. N Filipović: writing – review & editing. K Nikolic: supervision, conceptualization, methodology, resources, writing – review & editing. M Crevar: supervision, conceptualization, methodology, formal analysis, writing – review & editing.
Financial disclosure
This research was funded by the Ministry of Science, Technological Development and Innovation, Republic of Serbia through Grant Agreement with University of Belgrade – Faculty of Pharmacy No: 451-03-66/2024-03/200161, Grant Agreement with University of Belgrade – “VINČA” Institute of Nuclear Sciences – National Institute of the Republic of Serbia No: 451-03-66/2024-03/200017 and Institute for Medicinal Plant Research “Dr. Josif Pančić” No: 451-03-66/2024-03/200003. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Competing interests disclosures
The authors have no competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Writing disclosure
No writing assistance was utilized in the production of this manuscript.
Acknowledgments
We are grateful to research groups of V Savic from Faculty of Pharmacy, University of Belgrade, N Filipović from Faculty of Agriculture, University of Belgrade and T Todorović from Faculty of Chemistry, University of Belgrade for kindly providing us with experimental compounds used in this study.

