Figures & data
Figure 1. Schematic view of site-selective scission of double-stranded DNA by the first-generation ARCUT (combination of a pair of pcPNA + Ce(IV)-EDTA).
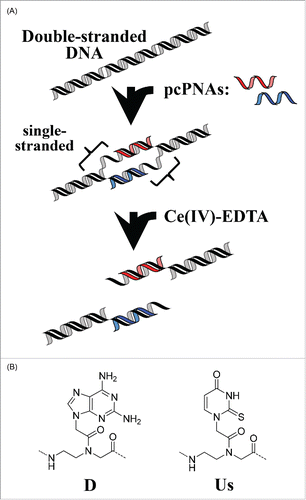
Figure 2. (A) Chemical structure of NLS-pcPNA conjugate. (B) Sequences of NLS-pcPNAs and pcPNAs. The NLS peptide was directly attached to the C-terminus of pcPNA, and all the pcPNAs have a phosphoserine (pS) at the N-termini. The 2-thiouracil Us is shown as U here.
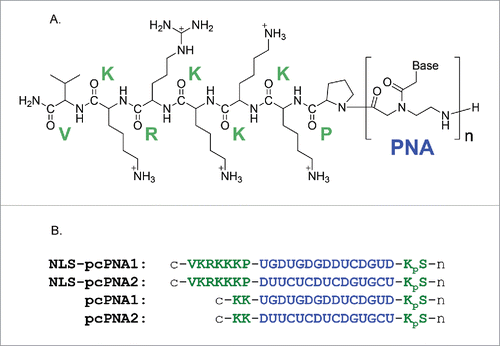
Figure 3. Gel mobility shift assay for the formation of invasion complex using pcPNAs with or without the conjugation of NLS peptide. Invasion conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 100 or 150 nM and [Hepes (pH 7.0)] = 5 mM at 50°C for 1 h. Reproduced by permission from ref. 37.
![Figure 3. Gel mobility shift assay for the formation of invasion complex using pcPNAs with or without the conjugation of NLS peptide. Invasion conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 100 or 150 nM and [Hepes (pH 7.0)] = 5 mM at 50°C for 1 h. Reproduced by permission from ref. 37.](/cms/asset/962a3544-4f03-440b-9d28-b42fece311a9/kdpx_a_1112457_f0003_oc.gif)
Figure 4. Invasion complex formation under intracellular salt conditions. [double-stranded DNA (119 bp)] = 50 nM, [NaCl] = 12 mM, [KCl] = 140 mM, and [MgCl2] = 0.8 mM at pH 7.0 and 50°C for 2 h. Reproduced by permission from ref. 37.
![Figure 4. Invasion complex formation under intracellular salt conditions. [double-stranded DNA (119 bp)] = 50 nM, [NaCl] = 12 mM, [KCl] = 140 mM, and [MgCl2] = 0.8 mM at pH 7.0 and 50°C for 2 h. Reproduced by permission from ref. 37.](/cms/asset/d4d55fa5-7315-4e35-89d1-0276de693a3f/kdpx_a_1112457_f0004_oc.gif)
Figure 5. Time-courses of invasion-complex formation. Conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 150 nM, [Hepes (pH 7.0)] = 5 mM and [NaCl] = 100 mM at 50°C. Reproduced by permission from ref. 37.
![Figure 5. Time-courses of invasion-complex formation. Conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 150 nM, [Hepes (pH 7.0)] = 5 mM and [NaCl] = 100 mM at 50°C. Reproduced by permission from ref. 37.](/cms/asset/82c29f52-8c35-4d83-abf9-c5f2a354a30f/kdpx_a_1112457_f0005_oc.gif)
Figure 6. Site-selective scission of double-stranded DNA by artificial DNA cutters composed of NLS-pcPNAs at high NaCl concentration (100 mM) at 50°C for 16 h. The scission mixtures involve [DNA substrate (4,733 bp)] = 4 nM, [HEPES (pH 7.0)] = 5 mM, and [Ce(IV)-EDTA] = 100 μM. The invasion complexes were prepared by incubating the mixtures at 50°C for 2 h. Reproduced by permission from ref. 37.
![Figure 6. Site-selective scission of double-stranded DNA by artificial DNA cutters composed of NLS-pcPNAs at high NaCl concentration (100 mM) at 50°C for 16 h. The scission mixtures involve [DNA substrate (4,733 bp)] = 4 nM, [HEPES (pH 7.0)] = 5 mM, and [Ce(IV)-EDTA] = 100 μM. The invasion complexes were prepared by incubating the mixtures at 50°C for 2 h. Reproduced by permission from ref. 37.](/cms/asset/a15ca45b-5526-432c-b41a-d41ada8f516a/kdpx_a_1112457_f0006_oc.gif)
Figure 7. (A) Recognition of double-stranded DNA by combining the invasion of 2 pcPNA strands (decamers) and Py-Im polyamide binding (recognizing 6 bp in DNA). (B) Conjugate of decamer pcPNA and Py-Im polyamide. DNA sequences recognized by each portion are presented. Reproduced by permission from ref. 49.
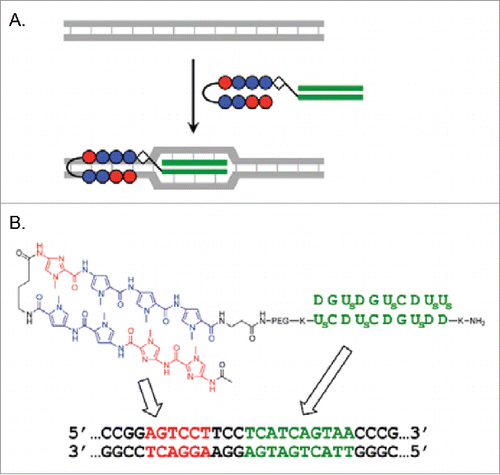
Figure 8. (A) Binding of DNA (130 bp) by the combination of the pcPNA3/Py-Im conjugate and pcPNA4 at higher salt conditions. Lane 1, DNA only; lanes 2–4, with pcPNA3/pcPNA4 (without the polyamide); lanes 5–7, combination of the conjugate and pcPNA4; M, 100 bp ladder. Invasion conditions: [DNA] = 10 nM, [conjugate] = [pcPNA3] = [pcPNA4] = 100 nM, [HEPES] = 5 mM at pH 7.0 and 37°C for 20 h. (B) Gel-shift assay for DNA-binding under physiological salt conditions ([NaCl] = 12 mM, [KCl] = 139 mM and [MgCl2] = 0.8 mM). Lane 1, DNA only; lane 2, with pcPNA3/pcPNA4 combination (200 nM each); lane 3, with the conjugate/pcPNA4 combination (200 nM each). Invasion conditions: [DNA] = 10 nM at pH 7.0 and 37°C for 20 h. Reproduced by permission from ref. 49.
![Figure 8. (A) Binding of DNA (130 bp) by the combination of the pcPNA3/Py-Im conjugate and pcPNA4 at higher salt conditions. Lane 1, DNA only; lanes 2–4, with pcPNA3/pcPNA4 (without the polyamide); lanes 5–7, combination of the conjugate and pcPNA4; M, 100 bp ladder. Invasion conditions: [DNA] = 10 nM, [conjugate] = [pcPNA3] = [pcPNA4] = 100 nM, [HEPES] = 5 mM at pH 7.0 and 37°C for 20 h. (B) Gel-shift assay for DNA-binding under physiological salt conditions ([NaCl] = 12 mM, [KCl] = 139 mM and [MgCl2] = 0.8 mM). Lane 1, DNA only; lane 2, with pcPNA3/pcPNA4 combination (200 nM each); lane 3, with the conjugate/pcPNA4 combination (200 nM each). Invasion conditions: [DNA] = 10 nM at pH 7.0 and 37°C for 20 h. Reproduced by permission from ref. 49.](/cms/asset/efe212bd-e18f-40db-bf89-ac6b3df2e05b/kdpx_a_1112457_f0008_oc.gif)
Figure 9. Site-selective hydrolysis of double-stranded DNA (4366 bp) with the pcPNA3/Py-Im conjugate and pcPNA4 combination. Lane 1, control; lane 2, with Ce(IV)-EDTA only; lane 3, with Ce(IV)-EDTA in the presence of the combination. Reaction conditions: [DNA] = 4 nM, [conjugate] = [pcPNA4] = 200 nM, [Ce(IV)-EDTA] = 200 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 49.
![Figure 9. Site-selective hydrolysis of double-stranded DNA (4366 bp) with the pcPNA3/Py-Im conjugate and pcPNA4 combination. Lane 1, control; lane 2, with Ce(IV)-EDTA only; lane 3, with Ce(IV)-EDTA in the presence of the combination. Reaction conditions: [DNA] = 4 nM, [conjugate] = [pcPNA4] = 200 nM, [Ce(IV)-EDTA] = 200 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 49.](/cms/asset/f06ab292-f6f0-447b-ad28-446479922f33/kdpx_a_1112457_f0009_oc.gif)
Figure 10. Site-selective scission of DNA by pcPNAs using the cutters prepared by in situ oxidation of Ce(III). (A) DNA scission with EDTA-bearing pcPNA. Lanes 1 and 2, pcPNA-5EDTA/pcPNA-6EDTA; Lanes 3 and 4, pcPNA-5/pcPNA-6. Lane M, 1,000 bp ladder. Reaction conditions: [DNA] = 4 nM, [each pcPNA] = 100 nM, [NaCl] = 100 mM at pH 7.0 and 50°C for 17 h under air. The conjugates were prepared using the monomer presented at the top of this figure. (B) Comparison of the effect of EDTA ligand with bisP ligand ([Ce(NO3)3] = 30 μM). Lane 1, DNA only; Lane 2, + Ce(III) only; Lane 3, pcPNA-5EDTA/pcPNA-6EDTA + Ce(III); Lane 4, pcPNA-5bisP/pcPNA-6bisP + Ce(III). In all the conjugates, the ligand was attached to the N-terminus of pcPNA. Reproduced from ref. 53.
![Figure 10. Site-selective scission of DNA by pcPNAs using the cutters prepared by in situ oxidation of Ce(III). (A) DNA scission with EDTA-bearing pcPNA. Lanes 1 and 2, pcPNA-5EDTA/pcPNA-6EDTA; Lanes 3 and 4, pcPNA-5/pcPNA-6. Lane M, 1,000 bp ladder. Reaction conditions: [DNA] = 4 nM, [each pcPNA] = 100 nM, [NaCl] = 100 mM at pH 7.0 and 50°C for 17 h under air. The conjugates were prepared using the monomer presented at the top of this figure. (B) Comparison of the effect of EDTA ligand with bisP ligand ([Ce(NO3)3] = 30 μM). Lane 1, DNA only; Lane 2, + Ce(III) only; Lane 3, pcPNA-5EDTA/pcPNA-6EDTA + Ce(III); Lane 4, pcPNA-5bisP/pcPNA-6bisP + Ce(III). In all the conjugates, the ligand was attached to the N-terminus of pcPNA. Reproduced from ref. 53.](/cms/asset/1f1cae25-d6d6-43a1-a140-c24a34eeed0c/kdpx_a_1112457_f0010_oc.gif)
Figure 11. Schematic representations of (i) site-selective DNA cutter using 2 pcPNA strands (the first-generation ARCUT) and (ii) new DNA cutter using only one strand of NLS-attached conventional PNA. The NLS was directly attached to the C-terminus of conventional PNA bearing no pseudo-complementary bases. Reproduced by permission from ref. 60.
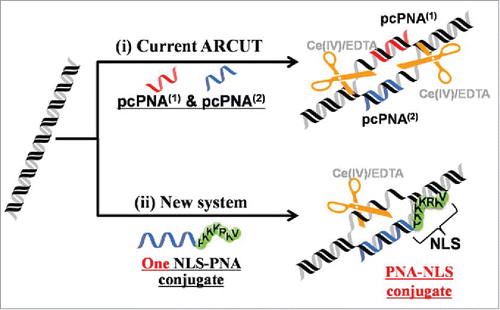
Figure 12. Site-selective DNA scission by combining one strand of PNA-NLS conjugate and Ce(IV)-EDTA. In (A) and (B), a 16-bp sequence in the exon 2 of MYCN gene and BFP gene in a plasmid was targeted, respectively. Lane 1, DNA only (control); Lane 2, DNA treated with Ce(IV)-EDTA (control); Lane 3, the scission with the combination of Ce(IV)-EDTA and PNA-NLS conjugate. Reaction conditions: [DNA] = 4.0 nM, [PNA-NLS] = 200 nM, [Ce(IV)-EDTA] = 200 µM, and [NaCl] = 100 mM at pH 7.0 and 37°C for 3 d The scission mixture was treated with Sma I (for (A)) or with Xba I (for (B)) before the electrophoresis. Reproduced by permission from ref. 60.
![Figure 12. Site-selective DNA scission by combining one strand of PNA-NLS conjugate and Ce(IV)-EDTA. In (A) and (B), a 16-bp sequence in the exon 2 of MYCN gene and BFP gene in a plasmid was targeted, respectively. Lane 1, DNA only (control); Lane 2, DNA treated with Ce(IV)-EDTA (control); Lane 3, the scission with the combination of Ce(IV)-EDTA and PNA-NLS conjugate. Reaction conditions: [DNA] = 4.0 nM, [PNA-NLS] = 200 nM, [Ce(IV)-EDTA] = 200 µM, and [NaCl] = 100 mM at pH 7.0 and 37°C for 3 d The scission mixture was treated with Sma I (for (A)) or with Xba I (for (B)) before the electrophoresis. Reproduced by permission from ref. 60.](/cms/asset/c9faa32c-f42d-45c7-acae-27f98a3af440/kdpx_a_1112457_f0012_oc.gif)
Figure 13. Fluorescence decay curves of the Quasar 570 dye-bound PNA2-NLS conjugate (excitation, 395 nm; photon counting, from 550 nm to 620 nm). Bottom, Dye-PNA2-NLS alone; top, Dye-PNA2-NLS + the plasmid. The values of τ1 (s), τ2 (s), A1, and A2, obtained by fitting the curves, are (0.14, 0.91, 0.82, 0.18) and (0.18, 1.72, 0.27, 0.73), respectively. For the purpose of comparison, the result for the 16-mer complementary oligonucleotide is shown by the middle curve. Measurement conditions are [Dye-PNA2-NLS] = 500 nM and [DNA] = 0 or 1000 nM at pH 7.0. Reproduced by permission from ref. 60.
![Figure 13. Fluorescence decay curves of the Quasar 570 dye-bound PNA2-NLS conjugate (excitation, 395 nm; photon counting, from 550 nm to 620 nm). Bottom, Dye-PNA2-NLS alone; top, Dye-PNA2-NLS + the plasmid. The values of τ1 (s), τ2 (s), A1, and A2, obtained by fitting the curves, are (0.14, 0.91, 0.82, 0.18) and (0.18, 1.72, 0.27, 0.73), respectively. For the purpose of comparison, the result for the 16-mer complementary oligonucleotide is shown by the middle curve. Measurement conditions are [Dye-PNA2-NLS] = 500 nM and [DNA] = 0 or 1000 nM at pH 7.0. Reproduced by permission from ref. 60.](/cms/asset/ca7ddbb0-86e4-4690-8c1b-c35fcd7d766b/kdpx_a_1112457_f0013_oc.gif)
Figure 14. Triple-helix forming bis-PNAs (A) and site-selective scission of double-stranded DNA by combining bis-PNA with Ce(IV)-EDTA (B). The single-stranded homopyrimidine sequence (the upper strand of DNA in (B) is hydrolyzed by the Ce(IV)-EDTA, and this primary scission promotes the hydrolysis of the counterpart homopurine sequence. The linker portion (PEG) is a consecutive linkage of 3 residues of 8-amino-3,6-dioxaoctanoic acid and K stands for a lysine residue. Reproduced by permission from ref. 66.
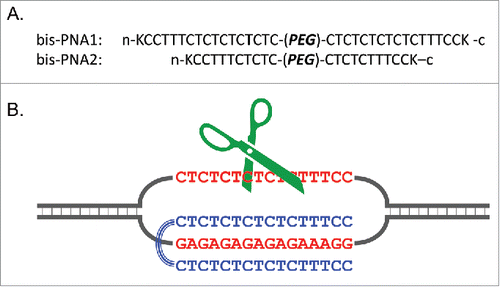
Figure 15. Site-selective scission of DNA (2685 bp) by combining triplex-forming bis-PNA1 and Ce(IV)-EDTA at the [bis-PNA1]/[DNA] ratios of 0 (lane 1), 10 (lane 2), and 50 (lane 3). Lane M, 500 bp ladder. Conditions; [DNA] = 4 nM, [bis-PNA1] = 40–200 nM, [Ce(IV)-EDTA] = 100 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 66.
![Figure 15. Site-selective scission of DNA (2685 bp) by combining triplex-forming bis-PNA1 and Ce(IV)-EDTA at the [bis-PNA1]/[DNA] ratios of 0 (lane 1), 10 (lane 2), and 50 (lane 3). Lane M, 500 bp ladder. Conditions; [DNA] = 4 nM, [bis-PNA1] = 40–200 nM, [Ce(IV)-EDTA] = 100 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 66.](/cms/asset/c7c21cf6-47ab-4103-958a-390cb9e94846/kdpx_a_1112457_f0015_b.gif)
Figure 16. Effects of molecular crowding conditions on the invasion complex formation of pcPNA with double-stranded DNA. [PEG200] = 0 (lanes 1 and 4), 20 (lanes 2 and 5), and 40 w/v % (lanes 3 and 6), respectively. The invasion conditions are [DNA (408-bp)] = 5 nM, [each pcPNA] = 100 nM, and [NaCl] = 100 mM at pH 7.0 (Hepes buffer) and 37°C for 1 h. In lanes 4–6, one base-pair in the DNA at the invasion site was changed from A/T to G/C, and one mismatch was introduced between the DNA strand and each of the pcPNAs. Lane M: 100 bp markers. Reproduced by permission from ref. 74.
![Figure 16. Effects of molecular crowding conditions on the invasion complex formation of pcPNA with double-stranded DNA. [PEG200] = 0 (lanes 1 and 4), 20 (lanes 2 and 5), and 40 w/v % (lanes 3 and 6), respectively. The invasion conditions are [DNA (408-bp)] = 5 nM, [each pcPNA] = 100 nM, and [NaCl] = 100 mM at pH 7.0 (Hepes buffer) and 37°C for 1 h. In lanes 4–6, one base-pair in the DNA at the invasion site was changed from A/T to G/C, and one mismatch was introduced between the DNA strand and each of the pcPNAs. Lane M: 100 bp markers. Reproduced by permission from ref. 74.](/cms/asset/7551b127-335c-458e-af84-19eba31fad58/kdpx_a_1112457_f0016_b.gif)
Figure 17. Invasion complex formation of pcPNA with double-stranded DNA in the medium mimicking the inside of living cells ([K+] = 140 mM, [Na+] = 10 mM, and [Mg2+] = 0.5 mM) under the molecular crowding conditions. [PEG200] = 0 (lane 1), 20 (lane 2), and 40 w/v % (lane 3). Other invasion conditions are the same as . Reproduced by permission from ref. 74.
![Figure 17. Invasion complex formation of pcPNA with double-stranded DNA in the medium mimicking the inside of living cells ([K+] = 140 mM, [Na+] = 10 mM, and [Mg2+] = 0.5 mM) under the molecular crowding conditions. [PEG200] = 0 (lane 1), 20 (lane 2), and 40 w/v % (lane 3). Other invasion conditions are the same as Fig. 16. Reproduced by permission from ref. 74.](/cms/asset/41e90c5c-5c22-41b7-8a13-e1c034ce2b43/kdpx_a_1112457_f0017_b.gif)
