ABSTRACT
Recently, completely chemistry-based tools for site-selective scission of DNA (ARCUT) have been prepared by combining 2 strands of pseudo-complementary PNA (pcPNA: site-selective activator) and a Ce(IV)-EDTA complex (molecular scissors). Its site-specificity is sufficient to cut the whole human genome at one predetermined site. In this first-generation ARCUT, however, there still remain several problems to be solved for wider applications. This review presents recent approaches to solve these problems. They are divided into (i) covalent modification of pcPNA with other functional groups and (ii) new strategies using conventional PNA, in place of pcPNA, as site-selective activator. Among various chemical modifications, conjugation with positively-charged nuclear localization signal peptide is especially effective. Furthermore, unimolecular activators, a single strand of which successfully activates the target site in DNA for site-selective scission, have been also developed. As the result of these modifications, the site-selective scission by Ce(IV)-EDTA was achieved promptly even under high salt conditions which are otherwise unfavourable for double-duplex invasion. Furthermore, it has been shown that “molecular crowding effect,” which characterizes the inside of living cells, enormously promotes the invasion, and thus the invasion seems to proceed effectively and spontaneously in the cells. Strong potential of pcPNA for further applications in vivo and in vitro has been confirmed.
Abbreviations
| ARCUT | = | artificial restriction DNA cutter |
| Boc | = | tert-butyloxycarbonyl |
| EDTA | = | ethylenediamine-N,N,N′,N′-tetraacetic acid |
| NLS | = | nuclear localization signal |
| pcPNA | = | pseudo-complementary peptide nucleic acid |
| PNA, | = | peptide nucleic acid |
| PEG | = | poly(ethylene glycol) |
| Py-Im polyamide | = | pyrrole-imidazole polyamide. |
Introduction
Man-made tools that can cut DNA selectively at predetermined sites have been nowadays attracting much interest.Citation1-3 Manipulation of huge genomes of higher animals and plants is one of the primary goals of these studies. Naturally occurring restriction enzymes are inappropriate for this purpose, because (i) their site-specificity is too low to cut the genomes at one site and (ii) their recognition sites are limited in number and targetable sequence. We have already developed a completely chemistry-based tool for site-selective scission of DNA.Citation4,5 This artificial restriction DNA cutter (ARCUT; ) is composed of 2 strands of pseudo-complementary PNA (pcPNA: site-selective activator) and Ce(IV)-EDTA complex (molecular scissors). This DNA analog is easily prepared by solid-phase synthesis,Citation6 and very strongly binds to cDNA through Watson-Crick base-pairing.Citation7 Furthermore, a pair of pcPNA strands can invade double-stranded DNA to form so-called double-duplex invasion complex.Citation8,9 This invasion is facilitated by using pseudo-complementary nucleobases (2,6-diaminopurine D and 2-thiouracil Us), in place of conventional A and T, and destabilizing the pcPNA/pcPNA duplex. The double-duplex invasion induces various effects on biological functions in vivo and in vitro.Citation10-12 In ARCUT, the binding sites of these pcPNA strands to the corresponding DNA strands are laterally shifted each other so that predetermined sites in both DNA strands remain single-stranded (see the middle in ). These single-stranded portions are selectively cut by the Ce(IV)-EDTA complex, which hydrolyzes only single-stranded DNA, resulting in the aimed site-selective scission.Citation13 The site-specificity of ARCUT can be easily modulated by changing the lengths and the sequences of the pcPNAs used, and be made high enough to cut the whole human genome (3 × 10Citation9 base-pairs) at one predetermined site.Citation14 Many applications of this tool have been already documented. As an example, the telomere at the end of each human chromosome (e.g., Xp/Yp) was selectively clipped and compared in the length.Citation15 The sub-telomere region of a chromosome was cut with the use of ARCUT, and the corresponding telomere was obtained. Although the telomeric repeats are exactly the same for all the chromosomes ((TTAGGG)n in the case of human beings), the sequence in this region is specific to each chromosome and can be targeted by ARCUT. By using another ARCUT, the telomere at the end of another chromosome (e.g., 11q) was clipped. It was found that the length of telomere is different from a chromosome to chromosome even in the same cells, indicating that the telomeres have still more complicated and important roles than proposed before. Furthermore, ARCUT has opened the way to completely restriction-enzyme-free DNA manipulation. The fragments obtained by the ARCUT scission at desired site can be directly combined each other or with other fragments with the use of DNA ligase. Even when there is no recognition site of naturally occurring enzyme, required recombinant DNA is straightforwardly obtainable.Citation4,5 The cutters are also applicable to gene editing in human cells by cutting the genome at designated site and promoting the desired homologous recombination.Citation16-18
Figure 1. Schematic view of site-selective scission of double-stranded DNA by the first-generation ARCUT (combination of a pair of pcPNA + Ce(IV)-EDTA).
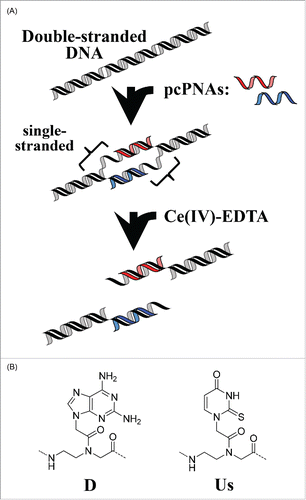
All these results confirm that ARCUT is a useful and highly promising tool for the future biology and biotechnology. In this first-generation ARCUT, however, there still remain several problems to be solved for even wider applications. The typical ones are as follows.
The sequences which are targetable should be still more flexible and versatile. Although they are rather freely chosen even with the first-generation ARCUT, some sites (e.g., highly GC rich regions) are difficult to target.
Site-selective scission by the ARCUT is not very effective in the media containing high concentrations of salts. This disadvantage must be solved especially for in vivo applications, since the inside of living cells is abundant in metal ions (K+, Na+, Mg2+, and others). It is well known that these high salt conditions are unfavorable for double-duplex invasion of unmodified pcPNAs (and thus for the site-selective scission by the ARCUT).
For in vivo applications, the cutters must be transferred to the nuclei and localized there where the genomes as target of site-selective scission exist.
At physiological temperatures, the invasion process is not sufficiently fast and cannot proceed spontaneously. In most of the previous experiments, the invasion complex was prepared by incubating the mixture at higher temperatures (e.g., 50°C).
It is more desirable if conventional (non pseudo-complementary) PNA can be used, in place of pcPNA. They are obtainable more easily and economically, and are directly relevant to naturally occurring DNA and RNA.
It has been already evidenced that both the site-selectivity and the scission efficiency of ARCUT are primarily governed by double-duplex invasion processes, in which the hot-spots for the scission (single-stranded portions) are formed in double-stranded DNA. In order to improve the functions of ARCUT and promote its applications in vivo and in vitro, these invasion processes must be contr-olled and promoted through covalent or non-covalent modification of pcPNA. This review presents recent approaches to solve these problems. Increased versatility of site-selective DNA cutters available should undoubtedly widen the scope of applications to biology, biotechnology, medicine, and others. At the same time, these improvements of the invading activity of pcPNA (and of PNA) should also greatly facilitate other applications of the invasion which have been already well documented by many laboratories.Citation19-36
Covalent Modifications of pcPNA to Enhance Site-Selective Scission by ARCUT
Conjugation of pcPNA with nuclear localization signal peptideCitation37
As described above, double-duplex invasion of (unmodified) pcPNA prefers low salt concentrations, and hardly occurs at physiological ionic strengths.Citation38-42 There, double-stranded DNA is so stable that the energy loss accompanied by its dissociation cannot be sufficiently compensated by the formation of 2 pcPNA/DNA duplexes in double-duplex invasion. The stability of these hetero-duplexes is not so much affected by ionic strength. One of the most plausible strategies to solve this problem is to attach positively-charged moieties to pcPNA and stabilize these duplexed through enhanced electrostatic interactions with negatively-charged DNA.Citation31 Based on these arguments, the nuclear localization signal (NLS) peptide of SV40 (PKKKRKV) was attached to the C-terminus of pcPNA strand (). This heptapeptide has large positive-charges (+5) at pH 7 and function in cells as “tag” to import a protein into the nuclei. Previously, conventional (non pseudo-complementary) PNA was modified with this oligopeptide. The conjugates were efficiently introduced to the cells, transferred to the nuclei, and exhibited antigene effects.Citation43-48
Figure 2. (A) Chemical structure of NLS-pcPNA conjugate. (B) Sequences of NLS-pcPNAs and pcPNAs. The NLS peptide was directly attached to the C-terminus of pcPNA, and all the pcPNAs have a phosphoserine (pS) at the N-termini. The 2-thiouracil Us is shown as U here.
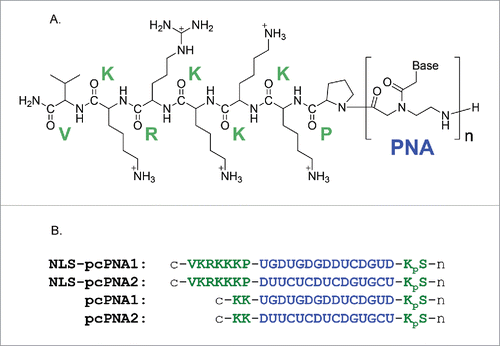
It was found that double-duplex invasion is greatly promoted by the conjugation of pcPNA with the NLS. The invasion of a pair of NLS-pcPNA conjugates efficiently occurred at small [conjugate]/[DNA] ratios where unmodified pcPNAs without the NLS hardly invaded the DNA (). Still more importantly, the invasion of the NLS-pcPNA is strongly resistant against ionic strength in the media. Even in the presence of 100 mM NaCl, where the invasion of unmodified pcPNA never occurred, this conjugate efficiently and promptly invaded the DNA. In , the salt concentrations were further increased up to the values in living cells: [NaCl] = 12 mM, [KCl] = 140 mM, and [MgCl2] = 0.8 mM. Consistently with the previous works from many laboratories, the invasion of a pair of unmodified pcPNAs was marginal under these conditions. With the NLS-pcPNA pair, however, notable invasion occurred even when [pcPNA]/[DNA] = 4 and became further dominant with the increase in the ratio. The mismatch-discrimination ability was also kept satisfactorily high for the NLS-pcPNA. Thus, the invasion completely disappeared, when one base-pair in 20-bp target DNA sequence was changed to another base-pair. By conjugating pcPNA with the NLS, double-duplex invasion has been successfully accomplished under the intracellular salt conditions. The electrostatic interactions between the positive charges of the NLS moiety and the negative charges of DNA simply strengthen the binding affinity of NLS-pcPNA to the target DNA, and satisfactorily compensate the energy loss due to the dissociation of the DNA/DNA duplex. Furthermore, the sequence-specificity in the invasion is hardly deteriorated in spite of the promoted electrostatic interactions. Superiority of the NLS is remarkable.
Figure 3. Gel mobility shift assay for the formation of invasion complex using pcPNAs with or without the conjugation of NLS peptide. Invasion conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 100 or 150 nM and [Hepes (pH 7.0)] = 5 mM at 50°C for 1 h. Reproduced by permission from ref. 37.
![Figure 3. Gel mobility shift assay for the formation of invasion complex using pcPNAs with or without the conjugation of NLS peptide. Invasion conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 100 or 150 nM and [Hepes (pH 7.0)] = 5 mM at 50°C for 1 h. Reproduced by permission from ref. 37.](/cms/asset/962a3544-4f03-440b-9d28-b42fece311a9/kdpx_a_1112457_f0003_oc.gif)
Figure 4. Invasion complex formation under intracellular salt conditions. [double-stranded DNA (119 bp)] = 50 nM, [NaCl] = 12 mM, [KCl] = 140 mM, and [MgCl2] = 0.8 mM at pH 7.0 and 50°C for 2 h. Reproduced by permission from ref. 37.
![Figure 4. Invasion complex formation under intracellular salt conditions. [double-stranded DNA (119 bp)] = 50 nM, [NaCl] = 12 mM, [KCl] = 140 mM, and [MgCl2] = 0.8 mM at pH 7.0 and 50°C for 2 h. Reproduced by permission from ref. 37.](/cms/asset/d4d55fa5-7315-4e35-89d1-0276de693a3f/kdpx_a_1112457_f0004_oc.gif)
In addition to the thermodynamic promotions described above, the NLS conjugation also increased the rate of invasion to significant extent. As shown in , the NLS-pcPNAs invaded the DNA promptly, and the reaction virtually completed only in about 30 min. The conversion in the plateau of the time-course (fraction of the invasion complex in the solution to the starting DNA) was around 90%. On the other hand, the invasion of unmodified pcPNAs was far slower and took more than 2 h to attain the plateau (the conversion = 35%). Thus, the NLS modification promoted the double-duplex invasion both thermodynamically and kinetically. Electrostatic attraction between the NLS and the DNA should be certainly one of the main factors responsible for these effects. However, this interaction cannot be the sole factor, since another positively-charged peptide RRRRR provides only poor results for the invasion and non-selective interactions of the conjugate with the DNA prevails there; vide infra. The primary structure of the NLS, its tertiary structure and/or its hydrophilicity/hydrophobicity balance should be also important.
Figure 5. Time-courses of invasion-complex formation. Conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 150 nM, [Hepes (pH 7.0)] = 5 mM and [NaCl] = 100 mM at 50°C. Reproduced by permission from ref. 37.
![Figure 5. Time-courses of invasion-complex formation. Conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of NLS-pcPNAs or pcPNAs] = 150 nM, [Hepes (pH 7.0)] = 5 mM and [NaCl] = 100 mM at 50°C. Reproduced by permission from ref. 37.](/cms/asset/82c29f52-8c35-4d83-abf9-c5f2a354a30f/kdpx_a_1112457_f0005_oc.gif)
In , the effect of the NLS on Ce(IV)-EDTA-mediated site-selective scission of DNA was examined under high salt conditions. First, the substrate DNA was treated with the combination of NLS-pcPNA1 and NLS-pcPNA2 in the presence of 100 mM NaCl, and then Ce(IV)-EDTA was added. Two scission products of expected sizes (1.7 and 3.0 kbp) were formed, confirming the site-selective scission. On the other hand, a pair of unmodified pcPNAs provided no scission products under the same conditions. These results are completely consistent with the invasion experiments presented above, and clearly show the essential role of the NLS moieties in the promotion of the site-selective DNA scission by ARCUT. Furthermore, the NLS moiety allows the conjugates to be transferred to the nuclei and preferentially localized there, promoting various in vivo applications.
Figure 6. Site-selective scission of double-stranded DNA by artificial DNA cutters composed of NLS-pcPNAs at high NaCl concentration (100 mM) at 50°C for 16 h. The scission mixtures involve [DNA substrate (4,733 bp)] = 4 nM, [HEPES (pH 7.0)] = 5 mM, and [Ce(IV)-EDTA] = 100 μM. The invasion complexes were prepared by incubating the mixtures at 50°C for 2 h. Reproduced by permission from ref. 37.
![Figure 6. Site-selective scission of double-stranded DNA by artificial DNA cutters composed of NLS-pcPNAs at high NaCl concentration (100 mM) at 50°C for 16 h. The scission mixtures involve [DNA substrate (4,733 bp)] = 4 nM, [HEPES (pH 7.0)] = 5 mM, and [Ce(IV)-EDTA] = 100 μM. The invasion complexes were prepared by incubating the mixtures at 50°C for 2 h. Reproduced by permission from ref. 37.](/cms/asset/a15ca45b-5526-432c-b41a-d41ada8f516a/kdpx_a_1112457_f0006_oc.gif)
Conjugation of pcPNA with pyrrole-imidazole polyamideCitation49
In another attempt to facilitate double-duplex invasion, pcPNA was conjugated with the pyrrole-imidazole polyamide (the balls in ). This Py-Im polyamide was developed by Dervan's group, and selectively binds to target site in double-stranded DNA.Citation50-52 The DNA binding of this polyamide is primarily based on multiple hydrogen bondings in the minor groove, and accompanies minimal structural change of the DNA. Thus, the binding successfully occurs even in the presence of high concentrations of salts. By taking advantage of this feature of Py-Im polyamide, double-duplex invasion of the conjugate and the resultant Ce(IV)/EDTA-mediated site-selective DNA cleavage were successfully accomplished under high salt conditions.
Figure 7. (A) Recognition of double-stranded DNA by combining the invasion of 2 pcPNA strands (decamers) and Py-Im polyamide binding (recognizing 6 bp in DNA). (B) Conjugate of decamer pcPNA and Py-Im polyamide. DNA sequences recognized by each portion are presented. Reproduced by permission from ref. 49.
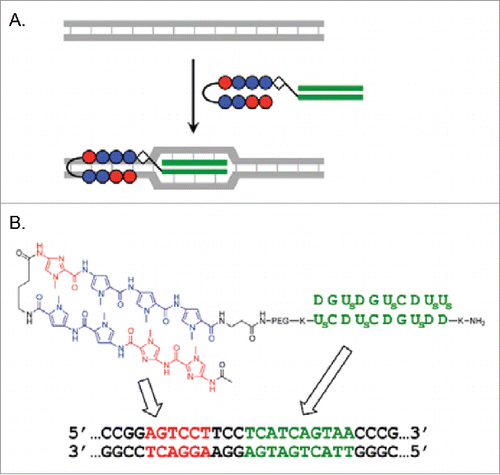
The conjugate of pcPNA3 and a Py-Im polyamide was synthesized using solid-phase synthesis. The pcPNA3 (H2N-DDUsGDCUsDCUs-COOH) is complementary to one strand of DNA (130 bp), whereas the Py-Im polyamide is designed to selectively bind to the 5′-AGTCCT-3′/3′-TCAGGA-5′ site in this DNA which is adjacent to the double-duplex invasion site of pcPNA3/pcPNA4. Accordingly, the combination of the conjugate and pcPNA4 (H2N-K2DGUsDGUsCDUsUsK-H) should recognize 10 bp by the double-duplex invasion and additionally the adjacent 5 bp by the minor groove binding. Importantly, the combination efficiently bound to the DNA even in the presence of 50–150 mM KCl (). In contrast, the binding of pcPNA3/pcPNA4 pair (without the conjugation with the Py-Im) was far weaker and greatly suppressed by the salts. The decameric pcPNAs are too short to form double-duplex invasion complexes efficiently under these conditions, and thus the strong resistance of the DNA binding by the Py-Im polyamide against metal salts is crucial here. The superior DNA binding capacity of the conjugate/pcPNA4 combination over the pcPNA3/pcPNA4 pair was also evident, when the KCl was replaced with MgCl2. In the presence of 1–2 mM MgCl2, most of the DNA formed the invasion complex with the combination, whereas only small portion bound to the pcPNA3/pcPNA4 pair. The mismatch recognition of the combination was also satisfactory. Upon the change of one C/G base-pair in the polyamide-binding site to A/T, the dissociation constant KD decreased by 30 fold according to surface plasmon resonance analysis. The conjugate bound to the fully-matching DNA much faster (ka = 7.8 × 103 M−1s−1) than the mismatching DNA (ka = 8.6 × 102 M−1s−1). When the number of base-pairs between the pcPNA-binding site and the polyamide-binding site was changed, the binding efficiency was as follows: CC < TCC > GTTCC. The optimal distance was 3 base-pairs.
Figure 8. (A) Binding of DNA (130 bp) by the combination of the pcPNA3/Py-Im conjugate and pcPNA4 at higher salt conditions. Lane 1, DNA only; lanes 2–4, with pcPNA3/pcPNA4 (without the polyamide); lanes 5–7, combination of the conjugate and pcPNA4; M, 100 bp ladder. Invasion conditions: [DNA] = 10 nM, [conjugate] = [pcPNA3] = [pcPNA4] = 100 nM, [HEPES] = 5 mM at pH 7.0 and 37°C for 20 h. (B) Gel-shift assay for DNA-binding under physiological salt conditions ([NaCl] = 12 mM, [KCl] = 139 mM and [MgCl2] = 0.8 mM). Lane 1, DNA only; lane 2, with pcPNA3/pcPNA4 combination (200 nM each); lane 3, with the conjugate/pcPNA4 combination (200 nM each). Invasion conditions: [DNA] = 10 nM at pH 7.0 and 37°C for 20 h. Reproduced by permission from ref. 49.
![Figure 8. (A) Binding of DNA (130 bp) by the combination of the pcPNA3/Py-Im conjugate and pcPNA4 at higher salt conditions. Lane 1, DNA only; lanes 2–4, with pcPNA3/pcPNA4 (without the polyamide); lanes 5–7, combination of the conjugate and pcPNA4; M, 100 bp ladder. Invasion conditions: [DNA] = 10 nM, [conjugate] = [pcPNA3] = [pcPNA4] = 100 nM, [HEPES] = 5 mM at pH 7.0 and 37°C for 20 h. (B) Gel-shift assay for DNA-binding under physiological salt conditions ([NaCl] = 12 mM, [KCl] = 139 mM and [MgCl2] = 0.8 mM). Lane 1, DNA only; lane 2, with pcPNA3/pcPNA4 combination (200 nM each); lane 3, with the conjugate/pcPNA4 combination (200 nM each). Invasion conditions: [DNA] = 10 nM at pH 7.0 and 37°C for 20 h. Reproduced by permission from ref. 49.](/cms/asset/efe212bd-e18f-40db-bf89-ac6b3df2e05b/kdpx_a_1112457_f0008_oc.gif)
It is noteworthy that the conjugate/pcPNA4 combination successfully formed invasion complex even under intracellular salt conditions in living cells ([NaCl] = 12 mM, [KCl] = 139 mM, and [MgCl2] = 0.8 mM). As shown in , the invasion of this combination to the DNA was notable even under these high salt conditions in which the pcPNA3/pcPNA4 pair without the NLS modification never showed the binding. Consistently, double-stranded DNA (4,366 bp) was site-selectively hydrolyzed by combining the Py-Im polyamide-pcPNA3 conjugate with pcPNA4 and treating the system with Ce(IV)-EDTA (). Two fragments (about 1,840 and 2,530 bp) for the scission at the binding site of the combination of the conjugate and pcPNA2 were produced. The hot spots for the scission (single-stranded portions) should be formed at the invasion-site, probably through enhanced molecular breathing motion. As expected, no scission occurred in the absence of the combination.
Figure 9. Site-selective hydrolysis of double-stranded DNA (4366 bp) with the pcPNA3/Py-Im conjugate and pcPNA4 combination. Lane 1, control; lane 2, with Ce(IV)-EDTA only; lane 3, with Ce(IV)-EDTA in the presence of the combination. Reaction conditions: [DNA] = 4 nM, [conjugate] = [pcPNA4] = 200 nM, [Ce(IV)-EDTA] = 200 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 49.
![Figure 9. Site-selective hydrolysis of double-stranded DNA (4366 bp) with the pcPNA3/Py-Im conjugate and pcPNA4 combination. Lane 1, control; lane 2, with Ce(IV)-EDTA only; lane 3, with Ce(IV)-EDTA in the presence of the combination. Reaction conditions: [DNA] = 4 nM, [conjugate] = [pcPNA4] = 200 nM, [Ce(IV)-EDTA] = 200 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 49.](/cms/asset/f06ab292-f6f0-447b-ad28-446479922f33/kdpx_a_1112457_f0009_oc.gif)
Attachment of metal-binding ligands to pcPNA and in situ preparation of site-selective DNA cutterCitation53
An EDTA was attached to the N-terminus of pcPNA with the use of a new Boc-monomer bearing tert-butyl-protected EDTA (the top in ), and site-selective cutter of double-stranded DNA was prepared by in situ oxidation of EDTA-bound Ce(III) to catalytically active Ce(IV). The EDTA-pcPNA conjugates were obtained by the solid-phase synthesis, and a pair of them was incubated with DNA substrate to form the double-duplex invasion complex. Then, CeIII(NO3)3 was added to the solution, and the Ce(III) was allowed to coordinate to the EDTA ligand. The DNA hydrolysis was carried out simply by incubating the mixture at 50°C under air. Although Ce(III) itself (its EDTA complex also) is virtually inactive for DNA hydrolysis, it was in situ converted to Ce(IV) through the oxidation by molecular O2, and acquired the eminent catalytic activity for DNA hydrolysis.Citation54-58 This in situ preparation of site-selective DNA cutter was experimentally confirmed by the results in . Only the DNA cutter, prepared from Ce(NO3)3 and a pair of EDTA-pcPNA conjugates (not from Ce(NO3)3 + a pair of pcPNA strands without EDTA), showed site-selective hydrolysis. In the site-selective DNA cutter prepared with this methodology, catalytically active Ce(IV) is bound to the EDTA and strictly located there, and thus its effective concentration for the catalysis is sufficiently large. Accordingly, the site-selective DNA scission can be still more clear-cut than the first-generation ARCUT, in which the site-selectivity for the scission is based on the difference in the activity for DNA hydrolysis (single-stranded DNA vs. double-stranded DNA). The pcPNA bearing an iminodi(methylphosphonic acid) (bisP), in place of EDTA, was also effective for this in situ preparation of site-selective DNA cutter ().
Figure 10. Site-selective scission of DNA by pcPNAs using the cutters prepared by in situ oxidation of Ce(III). (A) DNA scission with EDTA-bearing pcPNA. Lanes 1 and 2, pcPNA-5EDTA/pcPNA-6EDTA; Lanes 3 and 4, pcPNA-5/pcPNA-6. Lane M, 1,000 bp ladder. Reaction conditions: [DNA] = 4 nM, [each pcPNA] = 100 nM, [NaCl] = 100 mM at pH 7.0 and 50°C for 17 h under air. The conjugates were prepared using the monomer presented at the top of this figure. (B) Comparison of the effect of EDTA ligand with bisP ligand ([Ce(NO3)3] = 30 μM). Lane 1, DNA only; Lane 2, + Ce(III) only; Lane 3, pcPNA-5EDTA/pcPNA-6EDTA + Ce(III); Lane 4, pcPNA-5bisP/pcPNA-6bisP + Ce(III). In all the conjugates, the ligand was attached to the N-terminus of pcPNA. Reproduced from ref. 53.
![Figure 10. Site-selective scission of DNA by pcPNAs using the cutters prepared by in situ oxidation of Ce(III). (A) DNA scission with EDTA-bearing pcPNA. Lanes 1 and 2, pcPNA-5EDTA/pcPNA-6EDTA; Lanes 3 and 4, pcPNA-5/pcPNA-6. Lane M, 1,000 bp ladder. Reaction conditions: [DNA] = 4 nM, [each pcPNA] = 100 nM, [NaCl] = 100 mM at pH 7.0 and 50°C for 17 h under air. The conjugates were prepared using the monomer presented at the top of this figure. (B) Comparison of the effect of EDTA ligand with bisP ligand ([Ce(NO3)3] = 30 μM). Lane 1, DNA only; Lane 2, + Ce(III) only; Lane 3, pcPNA-5EDTA/pcPNA-6EDTA + Ce(III); Lane 4, pcPNA-5bisP/pcPNA-6bisP + Ce(III). In all the conjugates, the ligand was attached to the N-terminus of pcPNA. Reproduced from ref. 53.](/cms/asset/1f1cae25-d6d6-43a1-a140-c24a34eeed0c/kdpx_a_1112457_f0010_oc.gif)
This new methodology is reminiscent of in situ preparation of DNA cutters which selectively hydrolyze single-stranded DNAs.Citation59 There, a multiphosphonate ligand (nitrilotris(methylenephophonate) or N,N,N´,N´-ethylenediamine tetra(methylenephosphonate)) was bound to the ends of oligonucleotides. Using two of these modified oligonucleotides, a gap-structure was formed at the targeted scission-site in single-stranded DNA substrate so that the ligands were placed at the gap-site. Then, Ce(NO3)3 was added to the solution and the reaction was carried out under air. In the reaction mixture, most of the Ce(III) ions were bound to the multiphosphonate ligand, and converted by in situ oxidation to catalytically active Ce(IV) in the complex. Thus, the amount of unbound Ce(IV) in the reaction mixture, which could lead to undesired off-target scissions, was minimized. As the result, the scission by these cutters was so strictly restricted to the gap-site that single-stranded DNAs, even of considerable length (e.g., 800 base-length), were cut at the designated site with a very high selectivity. The scission fragments were successfully used to prepare recombinant DNAs.
Note that, in all other works described in this review, the source of Ce(IV) is the tetravalent salt CeIV(NH4)2(NO3)6, rather than the trivalent salts (e.g., CeIII(NO3)3). Accordingly, the activation process by the oxidation with molecular oxygen is never required to obtain the DNA cutters, and the site-selective scission can be straightforwardly and successfully achieved even in anaerobic conditions such as the inside of cells.
Use of Conventional (Non Pseudo-Complementary) PNA as Site-Selective Activator in ARCUT
Site-selective activation of double-stranded DNA by one strand of the conjugate between conventional PNA and nuclear localization signal peptide (NLS)Citation60
Even with the first-generation ARCUT, selective-scission site can be rather freely chosen. But still GC-rich regions are not very easy to be targeted, since the promotion of double-duplex invasion by pseudo-complementary bases D and Us, used for naturally occurring bases A and T, is one of the primary factors for effective function of this cutter. These two modified bases sterically repel each other and destabilize the duplex between the 2 pcPNA strands which are intrinsically complementary with each other. On the other hand, they form stable base-pairs with the corresponding counterpart bases A and T in DNA. Thus, pcPNA/DNA duplexes are sufficiently stable, allowing the efficient formation of the invasion complex. However, appropriate pseudo-complementary bases for G and C, which strongly bind to natural bases C and G but hardly form a mutual base-pair, have not yet been fully developed.Citation61-63 Thus, in GC-rich regions of substrate DNA, there is no efficient way to destabilize the corresponding pcPNA/pcPNA duplex and promote the invasion. One of the direct solutions to this problem is to develop a methodology in which only one pcPNA strand can activate the targeted site in double-stranded DNA. Here, conventional PNA involving only A, G, C, and T nucleobases can be directly used, since pcPNA/pcPNA duplexes are never formed. In fact, single PNA strand notably invaded double-stranded DNA when target site had a special sequence (e.g., inverted repeats) and in addition positively-charged amino acids or oligopeptides were attached to the PNA.Citation31 Chemical modification of the backbone with gamma PNA, as well as of nucleobases with G-clamp nucleobase, was also effective for the purpose.Citation39,41
On the basis of these findings, it was recently found that single strand of the PNA-NLS conjugate () successfully activates the target site in plasmid DNA (importantly, conventional PNA involving no pseudo-complementary bases was used there). Under the conditions where an unmodified PNA strand never binds to the DNA, a strand of this conjugate notably invades the DNA. The hot spot, formed by this single-molecular activator, was selectively hydrolyzed by Ce(IV)-EDTA (). Targeted scission sequences can be rather freely chosen. First, a sequence (16 bp) in exon 2 of N-myc oncogene (MYCN) was incorporated into a plasmid DNA, and targeted by an NLS-PNA1/Ce(IV)-EDTA system (). The NLS was directly attached to the C-terminus of 16-mer PNA1, which is complementary to the sense-strand (T340-A355) of MYCN. It should be noted that PNA1 involves only conventional nucleobases and no pseudo-complementary bases are used. This target sequence is highly GC rich (7 consecutive G-C pairs) and almost random without inverted repeat. When the PNA1 binds to the sense-strand according to the Watson-Crick rule, the corresponding antisense-strand becomes single-stranded. The site-selective hydrolysis of DNA (3,563 bp) by Ce(IV)-EDTA was carried out at pH 7.0 and 37°C. The single-stranded portion, formed by the unimolecular invasion, was hydrolyzed, converting the form I DNA (supercoiled) to the form II (relaxed open-circular), and then to the form III (linear). In order to pin down the scission site, the reaction mixture was further digested with a restriction enzyme Sma I, and the whole products were analyzed by agarose gel electrophoresis. Only two scission bands were observed, confirming the site-selective scission. The sizes of these 2 bands were exactly identical to those (around 1.5 and 2.0 kbp) expected from the double digestion by the NLS-PNA1/Ce(IV)-EDTA system and Sma I. The universal nature of this single-molecular activation was further confirmed by the selective scission of another 16 bp sequence in blue fluorescence protein (BFP) (). The scission by the NLS-PNA2/Ce(IV)-EDTA system (followed by digestion with Xba I) provided 2 fragments corresponding to the expected dual scission. It was concluded that only one strand of NLS-PNA conjugate is sufficient to activate the target site in the plasmid DNA and induces the site-selective scission by Ce(IV)-EDTA. As was the case in the first-generation ARCUT,Citation64 mismatch-discrimination ability of this system was satisfactory, and the scission was completely gone upon the change of one base-pair in the targeted scission sequence to another. When a strand of unmodified PNA was used as the activator, in place of the corresponding NLS conjugate, no DNA scission occurred.
Figure 11. Schematic representations of (i) site-selective DNA cutter using 2 pcPNA strands (the first-generation ARCUT) and (ii) new DNA cutter using only one strand of NLS-attached conventional PNA. The NLS was directly attached to the C-terminus of conventional PNA bearing no pseudo-complementary bases. Reproduced by permission from ref. 60.
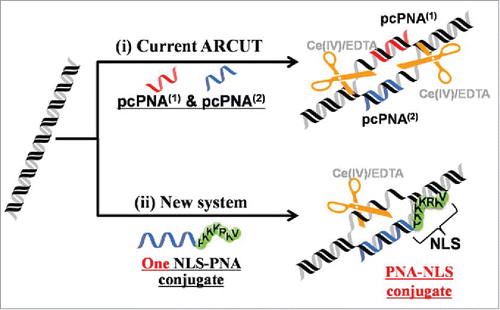
Figure 12. Site-selective DNA scission by combining one strand of PNA-NLS conjugate and Ce(IV)-EDTA. In (A) and (B), a 16-bp sequence in the exon 2 of MYCN gene and BFP gene in a plasmid was targeted, respectively. Lane 1, DNA only (control); Lane 2, DNA treated with Ce(IV)-EDTA (control); Lane 3, the scission with the combination of Ce(IV)-EDTA and PNA-NLS conjugate. Reaction conditions: [DNA] = 4.0 nM, [PNA-NLS] = 200 nM, [Ce(IV)-EDTA] = 200 µM, and [NaCl] = 100 mM at pH 7.0 and 37°C for 3 d The scission mixture was treated with Sma I (for (A)) or with Xba I (for (B)) before the electrophoresis. Reproduced by permission from ref. 60.
![Figure 12. Site-selective DNA scission by combining one strand of PNA-NLS conjugate and Ce(IV)-EDTA. In (A) and (B), a 16-bp sequence in the exon 2 of MYCN gene and BFP gene in a plasmid was targeted, respectively. Lane 1, DNA only (control); Lane 2, DNA treated with Ce(IV)-EDTA (control); Lane 3, the scission with the combination of Ce(IV)-EDTA and PNA-NLS conjugate. Reaction conditions: [DNA] = 4.0 nM, [PNA-NLS] = 200 nM, [Ce(IV)-EDTA] = 200 µM, and [NaCl] = 100 mM at pH 7.0 and 37°C for 3 d The scission mixture was treated with Sma I (for (A)) or with Xba I (for (B)) before the electrophoresis. Reproduced by permission from ref. 60.](/cms/asset/c9faa32c-f42d-45c7-acae-27f98a3af440/kdpx_a_1112457_f0012_oc.gif)
The critical importance of the NLS group for the present site-selective scission is definitely confirmed by the fact that the conjugate with another positively charged peptide (RRRRR) never functioned as unimolecular site-selective activator. When PNA-RRRRR conjugate was combined with Ce(IV)-EDTA, non-targeted sites were notably cut and the scission at the target site was never a dominant process. Apparently, this non-NLS conjugate bound to the DNA rather randomly through electrostatic interactions and activated various sites in this DNA. The primary structure of the NLS, its tertiary structure and/or the hydrophilicity/hydrophobicity balance would be important.
Invasion complex formation of the NLS-PNA conjugate with the target plasmid was further confirmed by fluorescence spectroscopy. A Quasar 570 dye was covalently bound to the N-terminus of NLS-PNA2 conjugate. The emission efficiency of this dye is primarily governed by the cis-trans isomerization in the photo-excited singlet state, and the quantum yield of this photo-isomerization sharply depends upon both the micro-environmental freedom and the interactions with adjacent aromatic residues.Citation65 It was found that the Dye-NLS-PNA2 conjugate emitted notable fluorescence only in the presence of the plasmid. The decay profiles were also remarkably affected by the plasmid (). All the decay curves were composed of 2 exponential terms, indicating the presence of 2 photo-excited singlet states. Without the plasmid, the shorter lifetime term (τ1 = 0.14 ns) was the major component (A1 = 82%, the bottom line). In its presence, however, the longer lifetime component (τ2 = 1.72 ns) became dominant (A2 = 73%, the top line). Upon the formation of invasion complex, the Quasar 570 dye is placed in a sterically restricted field and also in the vicinity of many nucleobases. Accordingly, its cis-trans photo-isomerization is notably suppressed, resulting in the increases in both the fluorescence intensity and the lifetime. Consistently, the complementary oligonucleotide showed smaller but similar effects on the fluorescence (the middle curve). Previously reported antigene effects of NLS-PNA conjugates for cancer treatments could be associated with this type of unimolecular invasion to the genome.Citation44-48
Figure 13. Fluorescence decay curves of the Quasar 570 dye-bound PNA2-NLS conjugate (excitation, 395 nm; photon counting, from 550 nm to 620 nm). Bottom, Dye-PNA2-NLS alone; top, Dye-PNA2-NLS + the plasmid. The values of τ1 (s), τ2 (s), A1, and A2, obtained by fitting the curves, are (0.14, 0.91, 0.82, 0.18) and (0.18, 1.72, 0.27, 0.73), respectively. For the purpose of comparison, the result for the 16-mer complementary oligonucleotide is shown by the middle curve. Measurement conditions are [Dye-PNA2-NLS] = 500 nM and [DNA] = 0 or 1000 nM at pH 7.0. Reproduced by permission from ref. 60.
![Figure 13. Fluorescence decay curves of the Quasar 570 dye-bound PNA2-NLS conjugate (excitation, 395 nm; photon counting, from 550 nm to 620 nm). Bottom, Dye-PNA2-NLS alone; top, Dye-PNA2-NLS + the plasmid. The values of τ1 (s), τ2 (s), A1, and A2, obtained by fitting the curves, are (0.14, 0.91, 0.82, 0.18) and (0.18, 1.72, 0.27, 0.73), respectively. For the purpose of comparison, the result for the 16-mer complementary oligonucleotide is shown by the middle curve. Measurement conditions are [Dye-PNA2-NLS] = 500 nM and [DNA] = 0 or 1000 nM at pH 7.0. Reproduced by permission from ref. 60.](/cms/asset/ca7ddbb0-86e4-4690-8c1b-c35fcd7d766b/kdpx_a_1112457_f0013_oc.gif)
As described in the section 1.1, a pair of the conjugates of pcPNA with the NLS efficiently invades double-stranded DNA. Thus, these conjugates show both double-duplex invasion and single-molecular invasion. Comprehensive study on the stability of these 2 kinds of invasion complexes and the other properties has not yet been achieved.
Site-selective DNA scission by combining a triplex-forming bis-PNA with Ce(IV)-EDTACitation66
Homopyrimidine/homopurine sequences take crucial roles in a range of cell functions (e.g., transcriptional regulation, chromatin organization, and DNA repair),Citation67,68 and are also proposed to be the binding sites of non-coding RNA for transcriptional regulation.Citation69-71 For the site-selective scission of these sites, the first-generation ARCUT is practically inapplicable, since pcPNA strands involving consecutive purine sequences are very difficult to prepare by solid-phase synthesis (in most cases, the coupling efficiency significantly decreases with increasing product length). However, these sequences were successfully targeted by combining triplex-formation of PNA with Ce(IV)-EDTA (). Two PNA strands were connected with appropriate linker to enhance the triplex-forming activity, and used as unimolecular site-selective activator (these kinds of conjugates are termed as bis-PNA).Citation72,73 Advantageously, only one PNA conjugate involving conventional PNA (containing no pseudo-complementary bases) is required to activate a predetermined site for the site-selective scission. Upon the triple-helix formation, single-stranded portion was formed in one DNA strand and selectively cut by Ce(IV)-EDTA. This primary scission in turn promoted the hydrolysis of the counterpart strand of the DNA, resulting in site-selective scission of both strands of the double-stranded DNA substrate.
Figure 14. Triple-helix forming bis-PNAs (A) and site-selective scission of double-stranded DNA by combining bis-PNA with Ce(IV)-EDTA (B). The single-stranded homopyrimidine sequence (the upper strand of DNA in (B) is hydrolyzed by the Ce(IV)-EDTA, and this primary scission promotes the hydrolysis of the counterpart homopurine sequence. The linker portion (PEG) is a consecutive linkage of 3 residues of 8-amino-3,6-dioxaoctanoic acid and K stands for a lysine residue. Reproduced by permission from ref. 66.
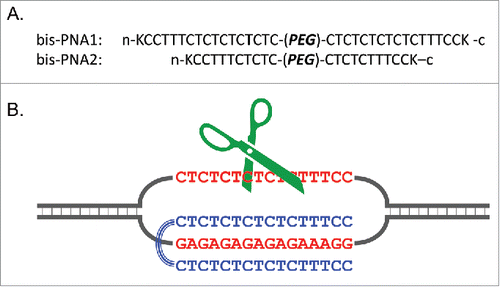
In , a 16 bp homopyrimidine/homopurine sequence in human chromosome 4 (5′-CTCTCTCTCTCTTTCC-3′/3′-GAGAGAGAGAGAAAGG-5′), being related to Hutchington's disease, was chosen as a target for the site-selective scission. In bis-PNA1, 2 homopyrimidine PNA strands (composed of 16 nucleobases) were connected by a poly(ethylene glycol) linker (PEG). The sequences of 2 PNA strands were designed on the basis of the formation of TAT and C+GC triplets. There is no need to use pseudo-complementary nucleobases, and thus conventional PNA can be employed. The combination of bis-PNA1 and Ce(IV)-EDTA successfully showed site-selective DNA scission of plasmid DNA which involved the target 16 bp sequence. When a 10 bp region (5′-CTCTCTTTCC-3′/3′-GAGAGAAAGG-5′) in plasmid was targeted with the use of a shorter bis-PNA2, site-selective DNA scission was also successful in terms of the same strategy. The sites which are difficult to target by the first-generation ARCUT have been successfully cut.
Figure 15. Site-selective scission of DNA (2685 bp) by combining triplex-forming bis-PNA1 and Ce(IV)-EDTA at the [bis-PNA1]/[DNA] ratios of 0 (lane 1), 10 (lane 2), and 50 (lane 3). Lane M, 500 bp ladder. Conditions; [DNA] = 4 nM, [bis-PNA1] = 40–200 nM, [Ce(IV)-EDTA] = 100 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 66.
![Figure 15. Site-selective scission of DNA (2685 bp) by combining triplex-forming bis-PNA1 and Ce(IV)-EDTA at the [bis-PNA1]/[DNA] ratios of 0 (lane 1), 10 (lane 2), and 50 (lane 3). Lane M, 500 bp ladder. Conditions; [DNA] = 4 nM, [bis-PNA1] = 40–200 nM, [Ce(IV)-EDTA] = 100 μM, and [NaCl] = 100 mM at pH 7.0 and 50°C for 16 h. Reproduced by permission from ref. 66.](/cms/asset/c7c21cf6-47ab-4103-958a-390cb9e94846/kdpx_a_1112457_f0015_b.gif)
Invasion Complex Formation Under Molecular Crowding ConditionsCitation74
In spite of a number of extracellular applications of pcPNA invasion, its intracellular applications have been rather limited, primarily because of the following 2 experimental results (note that they were obtained under non-molecular crowding conditions). First, the inside of cells is abundant in metal ions and thus seems to be inappropriate for the invasion. Second, the invasion is a rather slow process at physiological temperatures, and does not proceed spontaneously there (or takes a very long time for its completion). Although targeted mutation was successfully corrected with the use of a pair of pcPNAs in living cells,Citation75 details on the behaviors of these pcPNAs in the cells (whether they really formed the invasion complex or not) still remained unknown.
However, it has been confirmed that neither of these 2 factors appears to be critical for the applications in living cells, since “molecular crowding effect” Citation76,77 significantly facilitates the double-duplex invasion. The inside of cells is largely occupied by proteins and other biomolecules, and thus its physicochemical properties are much different from bulk aqueous phase. In the study, the influence of these specific intracellular environments (“molecular crowding effect”) was simulated by the addition of polyethylene glycol (PEG) to aqueous solutions.Citation78,79 It was found that PEG greatly promotes double-duplex invasion of pcPNA, and allows its formation at high salt conditions where otherwise the invasion never occurs. As an example, PEG of averaged molecular weight 200 (PEG200) was added to pH 7.0 Hepes buffer containing 100 mM NaCl (). The two pcPNA strands (H-K-UsUsDCUsGDUsGD-KK-NH2 and H-K-UsCDUsCDGUsDD-NH2) have 10 nucleobases. Without the addition of PEG200 ([each pcPNA] = 100 nM), the invasion never occurred at 37°C under the conditions employed. When PEG200 was added to this medium, however, the double-duplex invasion was remarkably promoted and notable amounts of invasion complexes were formed (lanes 2 and 3). Similar invasion-promoting effects were also observed for PEGs of other degrees of polymerization. When one mismatch was introduced between each pcPNA and its corresponding DNA strand by changing an A/T pair in the substrate DNA to G/C pair, no double-duplex invasion complex was detectable even in the presence of 20 % or 40 % (w/v) PEG200 (lanes 5 and 6). Mismatch-recognition under molecular crowding conditions was satisfactory. According to Sugimoto et al.,Citation80,81 both DNA/DNA duplex and PNA/DNA duplex are destabilized by molecular crowding effect, primarily due to the decrease of the activity of water in the media. However, the magnitude of destabilization of DNA/DNA duplex by this effect is far larger than the magnitude of destabilization of the corresponding destabilization of PNA/DNA duplex. As the result, double-duplex invasion is promoted by the molecular crowding effect. The destabilization of the DNA/DNA duplex under molecular crowding conditions should also facilitate the breathing motion of the duplex and be favorable for spontaneous formation of the invasion complex.
Figure 16. Effects of molecular crowding conditions on the invasion complex formation of pcPNA with double-stranded DNA. [PEG200] = 0 (lanes 1 and 4), 20 (lanes 2 and 5), and 40 w/v % (lanes 3 and 6), respectively. The invasion conditions are [DNA (408-bp)] = 5 nM, [each pcPNA] = 100 nM, and [NaCl] = 100 mM at pH 7.0 (Hepes buffer) and 37°C for 1 h. In lanes 4–6, one base-pair in the DNA at the invasion site was changed from A/T to G/C, and one mismatch was introduced between the DNA strand and each of the pcPNAs. Lane M: 100 bp markers. Reproduced by permission from ref. 74.
![Figure 16. Effects of molecular crowding conditions on the invasion complex formation of pcPNA with double-stranded DNA. [PEG200] = 0 (lanes 1 and 4), 20 (lanes 2 and 5), and 40 w/v % (lanes 3 and 6), respectively. The invasion conditions are [DNA (408-bp)] = 5 nM, [each pcPNA] = 100 nM, and [NaCl] = 100 mM at pH 7.0 (Hepes buffer) and 37°C for 1 h. In lanes 4–6, one base-pair in the DNA at the invasion site was changed from A/T to G/C, and one mismatch was introduced between the DNA strand and each of the pcPNAs. Lane M: 100 bp markers. Reproduced by permission from ref. 74.](/cms/asset/7551b127-335c-458e-af84-19eba31fad58/kdpx_a_1112457_f0016_b.gif)
Even under the high salt conditions in the inside of living cells ([Na+] = 10 mM, [K+] = 140 mM, and [Mg2+] = 0.5 mM), the invasion complex was formed to notable extent (). Importantly, the whole process of the double-duplex invasion spontaneously occurred at 37°C, and the rate was fast enough to complete the process at least within 1 h. This is highly in contrast with the fact that the double-duplex invasion under non-molecular crowding conditions was never a spontaneous process, and accordingly these complexes were usually prepared by incubating the components at higher temperatures (e.g., 50°C) for several hours. In these preparations, the salt concentrations in the media were in most cases kept minimal without adding any salts except for the buffer reagents. All these results substantiate that pcPNAs, incorporated into the living cells by some means, should easily bind to the target site in the genome through double-duplex invasion, and exhibit designated functions there.Citation10-12 Strong potential of pcPNA for further intracellular (and in vivo) applications has been confirmed.
Figure 17. Invasion complex formation of pcPNA with double-stranded DNA in the medium mimicking the inside of living cells ([K+] = 140 mM, [Na+] = 10 mM, and [Mg2+] = 0.5 mM) under the molecular crowding conditions. [PEG200] = 0 (lane 1), 20 (lane 2), and 40 w/v % (lane 3). Other invasion conditions are the same as . Reproduced by permission from ref. 74.
![Figure 17. Invasion complex formation of pcPNA with double-stranded DNA in the medium mimicking the inside of living cells ([K+] = 140 mM, [Na+] = 10 mM, and [Mg2+] = 0.5 mM) under the molecular crowding conditions. [PEG200] = 0 (lane 1), 20 (lane 2), and 40 w/v % (lane 3). Other invasion conditions are the same as Fig. 16. Reproduced by permission from ref. 74.](/cms/asset/41e90c5c-5c22-41b7-8a13-e1c034ce2b43/kdpx_a_1112457_f0017_b.gif)
Conclusions
Site-selective DNA cutter (ARCUT), previously developed by our group, is composed of 2 strands of pcPNA and Ce(IV)-EDTA complex.Citation4,5 The single-stranded portions, formed in both strands of double-stranded DNA through double-duplex invasion of the pcPNA, are hydrolyzed by Ce(IV)-EDTA. Recently, it has been shown that covalent and non-covalent modifications of pcPNA can greatly promote the double-duplex invasion and thus the resultant site-selective DNA scission. Some of the confronting problems of the first-generation ARCUT (the problems (1)-(4) presented in the introduction section) have been solved. Site-selective activation of double-stranded DNA by a single PNA strand has been also developed to provide new types of site-selective DNA cutters (solutions to the problems (1) and (5)). Furthermore, “molecular crowding effect,” which characterizes the specific environments in cells, has been shown to enormously facilitate the double-duplex invasion and allow successful invasion inside the living cells. It is indicated that the problems (2) and (4) are not critical when the pcPNA invasion is practically used in vivo for various applications. All these results show that double-duplex invasion of pcPNA (as well as ARCUT based on this invasion) should be highly promising for future applications in biochemistry, biology, medicine, and biotechnology.
In addition to these chemistry-based DNA cutters, protein-based DNA cutters have been also recently devised. Typical examples are conjugates of the nuclease domain of FokI restriction enzyme with either zinc finger proteins (ZFNCitation82) or transcription activator-like effectors (TALENCitation83,84). Homing endonucleases recognizing very long cognate sequences are engineered to target desired sites.Citation85 RNA-guided site-selective scission of genome by a nuclease (CRISPR-Cas system) has been also reported.Citation86,87 These cutters can be produced in living cells by transfecting the corresponding DNA vectors and used for intracellular gene editing. On the other hand, chemistry-based DNA-cutters have a remarkable feature that they can be freely modulated, according to our need, through chemical modification. Thus, required functions can be easily provided. This flexibility of molecular design is one of the most significant advantages of these cutters. Apparently, both chemistry-based DNA cutters and protein-based DNA cutters have pros and cons, and should be complementarily used for further developments of the relevant fields.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Chandra M, Sachdeva A, Silverman SK. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat Chem Biol 2009; 5:718-20; PMID:19684594; http://dx.doi.org/10.1038/nchembio.201
- Aiba Y, Sumaoka J, Komiyama M. Artificial DNA cutters for DNA manipulation and genome engineering. Chem Soc Rev 2011; 40:5657-68; PMID:21566825; http://dx.doi.org/10.1039/c1cs15039a
- Mancin F, Scrimin P, Tecilla P. Progress in artificial metallonucleases. Chem Commun 2012; 48:5545-59; PMID:22543403; http://dx.doi.org/10.1039/c2cc30952a
- Komiyama M, Aiba Y, Yamamoto Y, Sumaoka J. Artificial restriction DNA cutter for site-selective scission of double-stranded DNA with tunable scission site and specificity. Nat Protoc 2008; 3:655-62; PMID:18388948; http://dx.doi.org/10.1038/nprot.2008.7
- Katada H, Komiyama M. Artificial restriction DNA cutters as new tools for gene manipulation. ChemBioChem 2009; 10:1279-88; PMID:19396851; http://dx.doi.org/10.1002/cbic.200900040
- Komiyama M, Aiba Y, Ishizuka T, Sumaoka J. Solid-phase synthesis of pseudo-complementary peptide nucleic acids. Nat Protoc 2008; 3:646-54; PMID:18388947; http://dx.doi.org/10.1038/nprot.2008.6
- Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature 1993; 365:566-8; PMID:7692304; http://dx.doi.org/10.1038/365566a0
- Lohse J, Dahl O, Nielsen PE. Double duplex invasion by peptide nucleic acid: A general principle for sequence-specific targeting of double-stranded DNA. Proc Natl Acad Sci USA 1999; 96:11804-8; PMID:10518531; http://dx.doi.org/10.1073/pnas.96.21.11804
- Haaima G, Hansen HF, Christensen L, Dahl O, Nielsen PE. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucl Acids Res 1997; 25:4639-43; PMID:9358176; http://dx.doi.org/10.1093/nar/25.22.4639
- Izvolsky KI, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry 2000; 39:10908-13; PMID:10978178; http://dx.doi.org/10.1021/bi000675e
- Protozanova E, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Pseudocomplementary PNAs as selective modifiers of protein activity on duplex DNA: the case of type IIs restriction enzymes. Nucl Acids Res 2003; 31:3929-35; PMID:12853608; http://dx.doi.org/10.1093/nar/gkg450
- Kuhn H, Cherny DI, Demidov VV, Frank-Kamenetskii MD. Inducing and modulating anisotropic DNA bends by pseudocomplementary peptide nucleic acids. Proc Natl Acad Sci USA 2004; 101:7548-53; PMID:15136738[http://dx.doi.org/10.1073/pnas.0308756101
- Kitamura Y, Komiyama M. Preferential hydrolysis of gap and bulge sites in DNA by Ce(IV)/EDTA complex. Nucl Acids Res 2002; 30: e102; PMID:12364619; http://dx.doi.org/10.1093/nar/gnf101
- Ito K, Katada H, Shigi N, Komiyama M. Site-selective scission of human genome by artificial restriction DNA cutter. Chem Commun 2009; 6542-4; PMID:19865643; http://dx.doi.org/10.1039/b911208a
- Ishizuka T, Xu Y, Komiyama M. A chemistry-based method to detect individual telomere lengths at a single chromosome terminus. J Am Chem Soc 2013;135:14-7; PMID:23252341; http://dx.doi.org/10.1021/ja308481c
- Katada H, Chen HJ, Shigi N, Komiyama M. Homologous recombination in human cells using artificial restriction DNA cutter. Chem Commun 2009; 6545-7; PMID:19865644; http://dx.doi.org/10.1039/b912030k
- Katada H, Komiyama M. Artificial Restriction DNA Cutters to Promote Homologous Recombination in Human Cells. Curr Gene Ther 2011; 11:38-45; PMID:21182465; http://dx.doi.org/10.2174/156652311794520094
- Ito K, Shigi N, Komiyama M. Artificial restriction DNA cutter for site-selective gene insertion in human cells. Chem Commun 2013; 49:6764-6; PMID:23778429; http://dx.doi.org/10.1039/c3cc43261k
- Nielsen PE. Sequence-selective targeting of duplex DNA by peptide nucleic acids. Curr Opin Mol Ther 2010; 12:184-91; PMID:20373262
- Onyshchenko MI, Gaynutdinov TI, Englund EA, Appella DH, Neumann RD, Panyutin IG. Quadruplex formation is necessary for stable PNA invasion into duplex DNA of BCL2 promoter region. Nucl Acids Res 2011; 39:7114-23; PMID:21593130; http://dx.doi.org/10.1093/nar/gkr259
- Xu Y, Suzuki Y, Lönnberg T, Komiyama M. Human telomeric DNA sequence-specific cleaving by G-quadruplex formation. J Am Chem Soc 2009; 131:2871-4; PMID:19209856; http://dx.doi.org/10.1021/ja807313x
- Hu J, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA)-peptide conjugates that target chromosomal DNA. Biochemistry 2007; 46:7581-9; PMID:17536840; http://dx.doi.org/10.1021/bi700230a
- Bentin T, Hansen GI, Nielsen PE. Structural diversity of target-specific homopyrimidine peptide nucleic acid-dsDNA complexes. Nucl Acids Res 2006; 34:5790-9; PMID:17053099; http://dx.doi.org/10.1093/nar/gkl736
- Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA–DNA conjugates. Proc Natl Acad Sci USA 2002; 99:16695-700; PMID:12461167; http://dx.doi.org/10.1073/pnas.262556899
- Vickers TA, Griffith MC, Ramasamy K, Risen LM, Freier SM. Inhibition of NF-κB specific transcriptional activation by PNA strand invasion. Nucl Acids Res 1995; 23:3003-8; PMID:7659524; http://dx.doi.org/10.1093/nar/23.15.3003
- Demidov VV, Yavnilovich MV, Belotserkovskii BP, Frank-Kamenetskii MD, Nielsen PE. Kinetics and mechanism of polyamide (“peptide”) nucleic acid binding to duplex DNA. Proc Natl Acad Sci USA 1995; 92:2637-41; PMID:7708697; http://dx.doi.org/10.1073/pnas.92.7.2637
- Boffa LC, Carpaneto EM, Allfrey VG. Isolation of active genes containing CAG repeats by DNA strand invasion by a peptide nucleic acid. Proc Natl Acad Sci USA 1995; 92:1901-5; PMID:7892196; http://dx.doi.org/10.1073/pnas.92.6.1901
- Ackermann D, Famulok M. Pseudo-complementary PNA actuators as reversible switches in dynamic DNA nanotechnology. Nucl Acids Res 2013; 41:4729-39; PMID:23444144; http://dx.doi.org/10.1093/nar/gkt121
- Demidov VV, Protozanova E, Izvolsky KI, Price C, Nielsen PE, Frank-Kamenetskii MD. Kinetics and mechanism of the DNA double helix invasion by pseudocomplementary peptide nucleic acids. Proc Natl Acad Sci USA 2002; 99:5953-8; PMID:11972051; http://dx.doi.org/10.1073/pnas.092127999
- Datta B, Armitage BA. Hybridization of PNA to structured DNA targets: Quadruplex invasion and the overhang effect. J Am Chem Soc 2001; 123:9612-9; PMID:11572682; http://dx.doi.org/10.1021/ja016204c
- Zhang X, Ishihara T, Corey DR. Strand invasion by mixed base PNAs and a PNA-peptide chimera. Nucl Acids Res 2000; 28:3332-8; PMID:10954602; http://dx.doi.org/10.1093/nar/28.17.3332
- Ray A, Nordén B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J 2000; 14:1041-60; PMID:10834926
- Peffer NJ, Hanvey JC, Bisi JE, Thomson SA, Hassman CF, Noble SA, Babiss LE. Strand-invasion of duplex DNA by peptide nucleic acid oligomers. Proc Natl Acad Sci USA 1993; 90:10648-52; PMID:8248156; http://dx.doi.org/10.1073/pnas.90.22.10648
- Ishizuka T, Yoshida J, Yamamoto Y, Sumaoka J, Tedeschi T, Corradini R, Sforza S, Komiyama M. Chiral introduction of positive charges to PNA for double-duplex invasion to versatile sequences. Nucl Acids Res 2008; 36:1464-71; PMID:18203747; http://dx.doi.org/10.1093/nar/gkm1154
- Ishizuka T, Yang J, Komiyama M, Xu Y. G-rich sequence-specific recognition and scission of human genome by PNA/DNA hybrid G-quadruplex formation. Angew Chem Int Ed 2012; 51:7198-202; PMID:22700182; http://dx.doi.org/10.1002/anie.201201176
- Yamazaki T, Aiba Y, Yasuda K, Sakai Y, Yamanaka Y, Kuzuya A, Ohya Y, Komiyama M. Clear-cut observation of PNA invasion using nanomechanical DNA origami devices. Chem Commun 2012; 48:11361-3; PMID:23073563; http://dx.doi.org/10.1039/c2cc36358e
- Aiba Y, Honda Y, Komiyama M. Promotion of double-duplex invasion of peptide nucleic acid through conjugation with nuclear localization signal peptide. Chem Eur J 2015; 21:4021-6; PMID:25640012; http://dx.doi.org/10.1002/chem.201406085
- Nielsen PE, Christensen L. Strand displacement binding of a duplex-forming homopurine PNA to a homopyrimidine duplex DNA target. J Am Chem Soc 1996; 118:2287-8; PMID:19306309; http://dx.doi.org/10.1021/ja953125q
- He G, Rapireddy S, Bahal R, Sahu B, Ly DH. Strand invasion of extended, mixed-sequence B-DNA by γPNAs. J Am Chem Soc 2009; 131:12088-90; PMID:19663424; http://dx.doi.org/10.1021/ja900228j
- Smolina IV, Demidov VV, Soldatenkov VA, Chasovskikh SG, Frank-Kamenetskii MD. End invasion of peptide nucleic acids (PNAs) with mixed-base composition into linear DNA duplexes. Nucl Acids Res 2005; 33:e146; PMID:16204449; http://dx.doi.org/10.1093/nar/gni151
- Rapireddy S, He G, Roy S, Armitage BA, Ly DH. Strand invasion of mixed-sequence B-DNA by acridine-linked, γ-peptide nucleic acid (γ-PNA). J Am Chem Soc 2007; 129:15596-600; PMID:18027941; http://dx.doi.org/10.1021/ja074886j
- Chenna V, Rapireddy S, Sahu B, Ausin C, Pedroso E, Ly DH. A simple cytosine to G-clamp nucleobase substitution enables chiral γ-PNAs to invade mixed-sequence double-helical B-form DNA. ChemBioChem 2008; 9:2388-91; PMID:18816545; http://dx.doi.org/10.1002/cbic.200800441
- Branden LJ, Mohamed AJ, Smith CIE. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat Biotechnol 1999; 17:784-7; PMID:10429244[http://dx.doi.org/10.1038/11726
- Cutrona G, Carpaneto EM, Ulivi M, Roncella S, Landt O, Ferrarini M, Boffa LC. Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nat Biotechnol 2000; 18:300-3; PMID:10700145; http://dx.doi.org/10.1038/73745
- Cogoi S, Codognotto A, Rapozzi V, Meeuwenoord N, van der Marel G, Xodo LE. Transcription inhibition of oncogenic KRAS by a mutation-selective peptide nucleic acid conjugated to the PKKKRKV nuclear localization signal peptide. Biochemistry 2005; 44:10510-9; PMID:16060660; http://dx.doi.org/10.1021/bi0505215
- Tonelli R, Purgato S, Camerin, C, Fronza R, Bologna F, Alboresi S, Franzoni M, Corradini R, Sforza S, Faccini A, et al. Antigene peptide nucleic acid specifically inhibits MYCN expression in human neuroblastoma cells leading to cell growth inhibition and apoptosis. Mol Cancer Ther 2005; 4:779-86; PMID:15897242; http://dx.doi.org/10.1158/1535-7163.MCT-04-0213
- Tonelli R, McIntyre A, Camerin C, Walters ZS, Di Leo K, Selfe J, Purgato S, Missiaglia E, Tortori A, Renshaw J, et al. Antitumor activity of sustained N-Myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. Clin Cancer Res 2012; 18:796-807; PMID:22065083; http://dx.doi.org/10.1158/1078-0432.CCR-11-1981
- Faccini A, Tortori A, Tedeschi T, Sforza S, Tonelli R, Pession A, Corradini R, Marchelli R. Circular dichroism study of DNA binding by a potential anticancer peptide nucleic acid targeted against the MYCN oncogene. Chirality 2008; 20:494-500; PMID:17963203; http://dx.doi.org/10.1002/chir.20489
- Kameshima W, Ishizuka T, Minoshima M, Yamamoto M, Sugiyama H, Xu Y, Komiyama M. Conjugation of peptide nucleic acid with pyrrole–imidazole polyamide to specifically recognize and cleave DNA. Angew Chem Int Ed 2013; 125:13926-9; PMID:24155125; http://dx.doi.org/10.1002/ange.201305489
- Turner JM, Swalley SE, Baird EE, Dervan PB. Aliphatic/aromatic amino acid pairings for polyamide recognition in the minor groove of DNA. J Am Chem Soc 1998; 120:6219-26; http://dx.doi.org/10.1021/ja980147e
- Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol 2003; 13:284-99; PMID:12831879; http://dx.doi.org/10.1016/S0959-440X(03)00081-2
- Bando T, Sugiyama H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole−imidazole polyamides. Acc Chem Res 2006; 39:935-44; PMID:17176032; http://dx.doi.org/10.1021/ar030287f
- Aiba Y, Honda Y, Han Y, Komiyama M. Introduction of multiphosphonate ligand to peptide nucleic acid for metal ion conjugation. Artificial DNA: PNA & XNA 2012; 3:73-9; PMID:22772037; http://dx.doi.org/10.4161/adna.20727
- Matsumoto Y, Komiyama M. DNA hydrolysis by rare earth metal ions. Chem Exp 1992; 7:785-8
- Sumaoka J, Yashiro M, Komiyama M. Remarkably fast hydrolysis of 3′,5′-cyclic adenosine mono-phosphate by cerium(III) hydroxide cluster. Chem Commun 1992; 1707-8; http://dx.doi.org/10.1039/c39920001707
- Komiyama M, Kodama T, Takeda N, Sumaoka J, Shiiba T, Matsumoto Y, Yashiro M. Catalytically active species for the CeCl3-induced DNA hydrolysis. J Biochem 1994; 115:809-10; PMID:7961589
- Takasaki BK, Chin J. Cleavage of the phosphate diester backbone of DNA with cerium(III) and molecular oxygen. J Am Chem Soc 1994; 116:1121-2; http://dx.doi.org/10.1021/ja00082a040
- Komiyama M, Shiiba T, Kodama T, Takeda N, Sumaoka J, Yashiro M. DNA hydrolysis by cerium(IV) does not involve either molecular oxygen or hydrogen peroxide. Chem Lett 1994; 23:1025-8; http://dx.doi.org/10.1246/cl.1994.1025
- Lönnberg T, Aiba Y, Hamano Y, Miyajima Y, Sumaoka J, Komiyama M. Oxidation of an oligonucleotide-bound Ce(III)/multiphosphonate complex for site-selective DNA scission. Chem Eur J 2010; 16:855-9; PMID:19938010; http://dx.doi.org/10.1002/chem.200902169
- Aiba Y, Hamano Y, Kameshima W, Araki Y, Wada T, Accetta A, Sforza S, Corradini R, Marchelli R, Komiyama M. Conjugate of peptide nucleic acid and nuclear localization signal as single-molecular activator of target site in double-stranded DNA for site-selective scission. Org Biomol Chem 2013; 11:5233-8; PMID:23820872; http://dx.doi.org/10.1039/c3ob40947c
- Lahoud G, Timoshchuk V, Lebedev A, Arar K, Hou YM, Gamper H. Properties of pseudo-complementary DNA substituted with weakly pairing analogs of guanine or cytosine. Nucl Acids Res 2008; 36:6999-7008; PMID:18987000; http://dx.doi.org/10.1093/nar/gkn797
- Hoshika S, Chen F, Leal NA, Benner SA. Artificial genetic systems: Self-avoiding DNA in PCR and multiplexed PCR. Angew Chem Int Ed 2010; 49:5554-7; PMID:20586087; http://dx.doi.org/10.1002/anie.201001977
- Olsen AG, Dahl O, Petersen AB, Nielsen J, Nielsen PE. A novel pseudo-complementary PNA G-C base pair. Artificial DNA: PNA & XNA 2011; 2:32-6; PMID:21686250; http://dx.doi.org/10.4161/adna.2.1.15554
- Miyajima Y, Ishizuka T, Yamamoto Y, Sumaoka J, Komiyama M. Origin of high fidelity in target-sequence recognition by PNA-Ce(IV)/EDTA combination as site-selective DNA cutter. J Am Chem Soc 2009; 131:2657-62; PMID:19199631; http://dx.doi.org/10.1021/ja808290e
- Harvey BJ, Levitus M. Nucleobase-specific enhancement of Cy3 fluorescence. J Fluoresc 2009; 19:443-8; PMID:18972191; http://dx.doi.org/10.1007/s10895-008-0431-1
- Aiba Y, Yasuda K, Komiyama M. Site-selective scission of double-stranded DNA by combining a triplex-forming bis-PNA and Ce(IV)/EDTA. Chem Lett 2013; 42:1300-2; http://dx.doi.org/10.1246/cl.130650
- Buske FA, Mattick JS, Bailey TL. Potential in vivo roles of nucleic acid triple-helices. RNA Biol 2011; 8:427-39; PMID:21525785; http://dx.doi.org/10.4161/rna.8.3.14999
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447:932-40; PMID:17581576; http://dx.doi.org/10.1038/nature05977
- Dey I, Rath PC. A novel rat genomic simple repeat DNA with RNA-homology shows triplex (H-DNA)-like structure and tissue-specific RNA expression. Biochem Biophys Res Commun 2005; 327:276-86; PMID:15629459; http://dx.doi.org/10.1016/j.bbrc.2004.12.015
- Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA 2010; 107:12816-21; PMID:20616059; http://dx.doi.org/10.1073/pnas.1007580107
- Belotserkovskii BP, Neil AJ, Saleh SS, Shin JHS, Mirkin SM, Hanawalt PC. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucl Acids Res 2013; 41:1817-28; PMID:23275544; http://dx.doi.org/10.1093/nar/gks1333
- Egholm M, Christensen L, Deuholm KL, Buchardt O, Coull J, Nielsen PE. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucl Acids Res 1995; 23:217-22; PMID:7862524; http://dx.doi.org/10.1093/nar/23.2.217
- Griffith MC, Risen LM, Greig MJ, Lesnik EA, Sprankle KG, Griffey RH, Kiely JS, Freier SM. Single and bis peptide nucleic acids as triplexing agents: Binding and stoichiometry. J Am Chem Soc 1995; 117:831-2; http://dx.doi.org/10.1021/ja00107a033
- Sumaoka J, Komiyama M. Molecular crowding facilitates double-duplex invasion of pseudo-complementary peptide nucleic acid in high salt medium. Chem Lett 2014; 43:1581-3; http://dx.doi.org/10.1246/cl.140620
- Lonkar P, Kim KH, Kuan JY, Chin JY, Rogers FA, Knauert MP, Kole R, Nielsen PE, Glazer PM. Targeted correction of a thalassemia-associated b-globin mutation induced by pseudo-complementary peptide nucleic acids. Nucl Acids Res 2009; 37:3635-44; PMID:19364810; http://dx.doi.org/10.1093/nar/gkp217
- Fulton AB. How crowded is the cytoplasm? Cell 1982; 30:345-7; PMID:6754085; http://dx.doi.org/10.1016/0092-8674(82)90231-8
- Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol 1991; 222:599-620; PMID:1748995; http://dx.doi.org/10.1016/0022-2836(91)90499-V
- Miyoshi D, Sugimoto N. Molecular crowding effects on structure and stability of DNA. Biochimie 2008; 90:1040-51; PMID:18331845; http://dx.doi.org/10.1016/j.biochi.2008.02.009
- Markarian MZ, Schlenoff JB. Effect of molecular crowding and ionic strength on the isothermal hybridization of oligonucleotides. J Phys Chem B 2010; 114:10620-7; PMID:20701389; http://dx.doi.org/10.1021/jp103213w
- Miyoshi D, Nakamura K, Tateishi-Karimata H, Ohmichi T, Sugimoto N. Hydration of Watson−Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J Am Chem Soc 2009; 131:3522-31; PMID:19236045; http://dx.doi.org/10.1021/ja805972a
- Nakano S, Yamaguchi D, Tateishi-Karimata H, Miyoshi D, Sugimoto N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys J 2012; 102:2808-17; PMID:22735531; http://dx.doi.org/10.1016/j.bpj.2012.05.019
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 1996; 93:1156-60; PMID:8577732; http://dx.doi.org/10.1073/pnas.93.3.1156
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010; 186:757-61; PMID:20660643; http://dx.doi.org/10.1534/genetics.110.120717
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucl Acids Res 2011; 39:9283-93; PMID:21813459; http://dx.doi.org/10.1093/nar/gkr597
- Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys 2005; 38:49-95; PMID:16336743; http://dx.doi.org/10.1017/S0033583505004063
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819-23; PMID:23287718; http://dx.doi.org/10.1126/science.1231143
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; http://dx.doi.org/10.1126/science.1232033

