Figures & data

Figure 1. Geographical distribution of different Lassa virus (LASV) lineages in West Africa. LASV lineages are determined based on phylogenetic analysis of viral nucleotide or amino acid sequence variations and the geographical clustering and location where the viruses were discovered. Lineages I–III, and VI are circulating in Nigeria. The lineage IV is found in Guinea, Sierra Leone, and Liberia, and lineage V in southern Mali. The proposed lineage VII is found in Togo. Map made with an outline obtained from Africa – MapChart at the following web-site: https://mapchart.net/africa.html
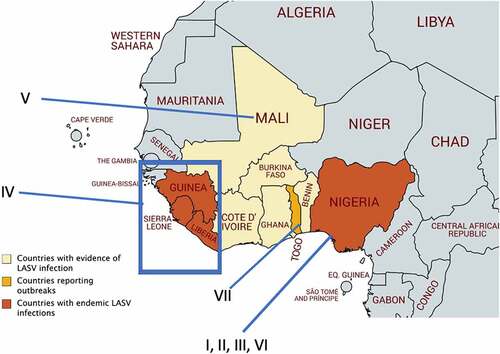
Figure 2. Mammarenaviral RNA genome structure, replication, transcription and gene expression strategies. (a) The bisegmented genome of arenaviruses (e.g., LASV) contains two genomic RNA segments that code for four known viral proteins in an ambisense coding strategy. Each genome segment contains two open reading frames that encode two gene products separated by a noncoding intergenic region (IGR) that forms stable hairpin RNA structure(s). The L segment is ~7.2 kb and codes for the L protein and the Z protein, while the S segment is ~3.5 kb and codes for the viral glycoprotein precursor (GPC) and NP proteins. (b) Mammarenaviral RNA replication, transcription and gene expression strategies. The L polymerase, together with NP, transcribes negative-sense genes (NP and L genes) starting at the 3ʹ untranslated region (UTR) toward the noncoding intergenic region (IGR) of the S genomic and L genomic RNAs in order to generate the viral L and NP mRNAs, respectively, from which the viral NP and L proteins are translated. Occasionally, the L polymerase, together with NP, continues RNA synthesis past the IGR to generate complementary S antigenomic RNA and L antigenomic RNA, which are used as templates to transcribe the GPC and Z mRNAs for translation into the respective viral proteins GPC and Z. GPC is post-translationally modified and processed by cellular proteases into GP1, GP2 and SSP protein subunits, which are incorporated into the cellular surface membrane where they interact with the viral Z protein and the viral ribonucleoprotein complex that consists of the viral NP, L and viral genomic RNAs for viral assembly and budding. The S and L antigenomic RNAs are used as templates by the viral L polymerase and NP to synthesize the full-length S and L genomic RNAs for incorporation into the viral RNPs for packaging into the newly formed virion particle
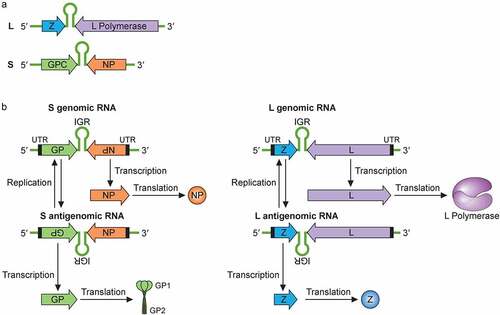
Figure 3. Mammrenavirus inhibition of innate immunity. The cellular Toll-like receptors (TLRs) recognize pathogen-associated molecular patterns (PAMPS) (e.g., viral glycoproteins) and utilize the cellular MYD88 adaptor protein to transmit a cellular signal to successively influence the activities of other cellular proteins, including TAK1, IKKα, in order to activate transcription factors, such as interferon regulatory factors (IRF3 and IRF7) and NFkB subunits to induce the expression of type I interferons (IFNα and IFNβ), which are secreted out of the cells to bind to their respective receptors on the surface membrane of the cells to further activate cellular protein kinases (TYK2, JAK1, JAK2) and transcription factors (STAT1 and STAT2 by phosphorylation and IRF9), which are translocated from the cellular cytoplasm into the nucleus to activate the expression of hundreds of antiviral genes from their respective promoters (GAS or ISRE). In addition to the TLRs, other intracellular receptors (e.g., RIG-I and MDA5) can recognize aberrant viral double-stranded RNAs (dsRNAs) as PAMPs to activate other cellular proteins (MAVS on the mitochrondria, DDX3, TBK, IKKe), which in turn, activate IRF3, IRF7 and NFkB transcription factors in order to upregulate the expression of type I interferon genes (IFNα and IFNβ). Aberrant virus-associated dsRNAs can also act as PAMPs to activate other intracellular receptor (PKR) to activate NFkB and influence the activity of the other cellular protein (e.g., eIF4E) in order to increase the expression of the type I interferon genes (IFNα and IFNβ). Mammarenaviral proteins, such as LASV NP and Z proteins, have been shown to use various strategies to inhibit the expression of type I interferon genes (IFNα and IFNβ)
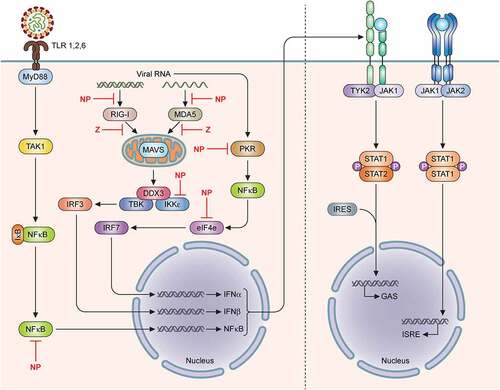
Table 1. Lassa vaccine candidates and platforms used in preclinical developments
Data availability statement
All figures and table included in this article have been deposited in a recognized data repository (Figshare.com) with a digital object identifier (10.6084/m9.figshare.14350274).
